Fast detection method of PCR amplification products of food pathogenic bacteria
A technology for amplification products and detection methods, which is applied in the field of rapid detection of PCR amplification products of pathogenic bacteria, can solve the problems of high detection cost of real-time fluorescent PCR method, small detection range of pathogenic bacteria, easy to cause pollution, etc., and achieve easy Popularization and application, low cost and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] A rapid detection method for PCR amplification products of Vibrio parahaemolyticus, the specific steps are as follows:
[0030] (1) Magnetic bead method to extract DNA template of Vibrio parahaemolyticus
[0031] A. Take 1mL of Vibrio parahaemolyticus, centrifuge at 10000r / min for 5min, remove the supernatant, dissolve the precipitate in 1mL of bacterial lysate, shake for 15s, in a water bath at 70℃ for 12min, centrifuge at 5000r / min to obtain 1mL of cell lysate; The formula of bacterial lysate is 50μL 1M Tris-HCl, 50μL 15wt% SDS solution, 50μL 20mg / ml proteinase K, 20μL 0.5M EDTA with pH=5.0, 2μL RNase, make up to 1mL with double distilled water;
[0032] B. Add 10 μL of magnetic nanoparticles to 1 mL of cell lysate, vortex for 5 seconds, then add 600 μL of adsorption buffer to mix, shake for 15 minutes, and magnetically separate the supernatant; the adsorption buffer consists of 80 μL of 9wt% polyethylene glycol Mixing 20000 solution and 500μL of 5M NaCl solution;
[0033]...
Embodiment 2
[0039] A rapid detection method for E. coli PCR amplification products, the specific steps are as follows:
[0040] (1) Extraction of E. coli DNA template by magnetic bead method
[0041] A. Take 1mL of Escherichia coli bacteria liquid, centrifuge at 10000r / min for 5min, remove the supernatant, dissolve the precipitate in 1mL of bacterial lysate, shake for 15s, 70℃ water bath for 12min, centrifuge at 5000r / min to obtain 1mL of cell lysate; where the bacteria are lysed The formulation of the solution is 50μL of 1M Tris-HCl, 50μL of 15wt% SDS solution, 50μL of 20mg / ml proteinase K, 20μL of 0.5M EDTA with pH=5.0, 2μL of RNase, and make up to 1mL with double distilled water;
[0042] B. Add 10 μL of magnetic nanoparticles to 1 mL of cell lysate, vortex for 5 seconds, then add 600 μL of adsorption buffer to mix, shake for 15 minutes, and magnetically separate the supernatant; the adsorption buffer consists of 80 μL of 9wt% polyethylene glycol Mixing 20000 solution and 500μL of 5M NaCl ...
Embodiment 3
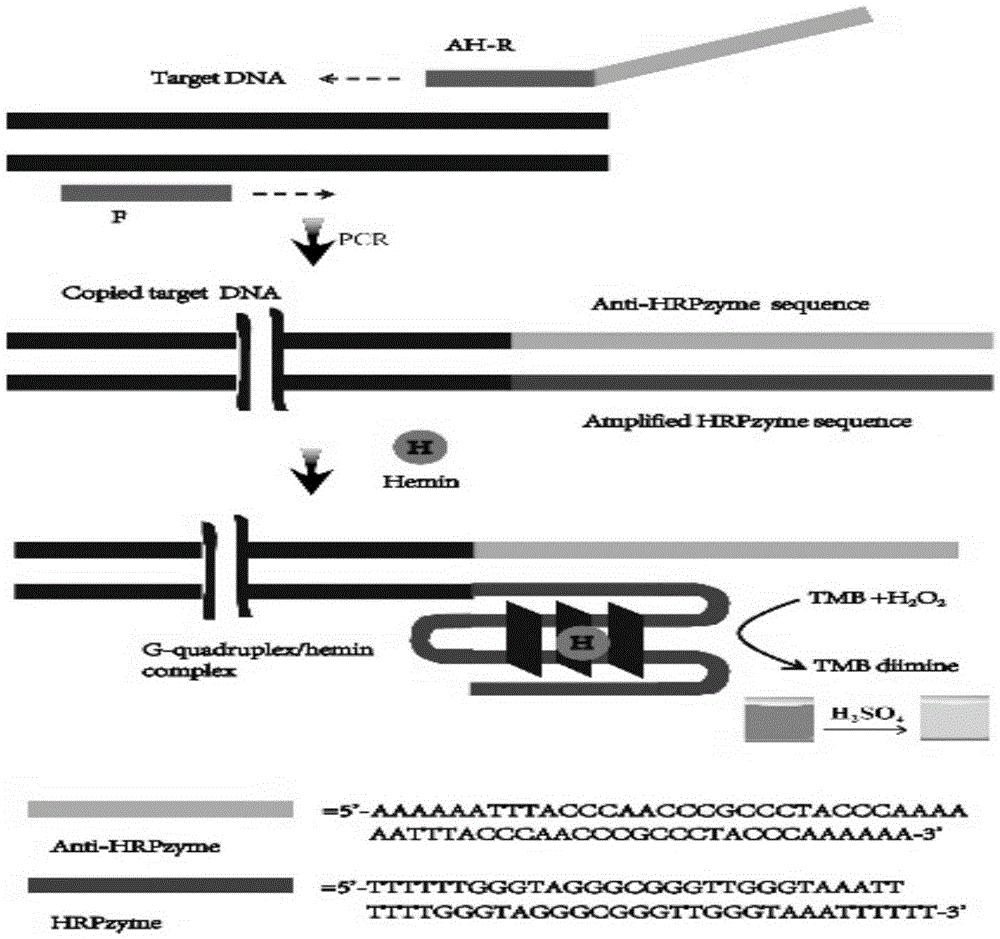
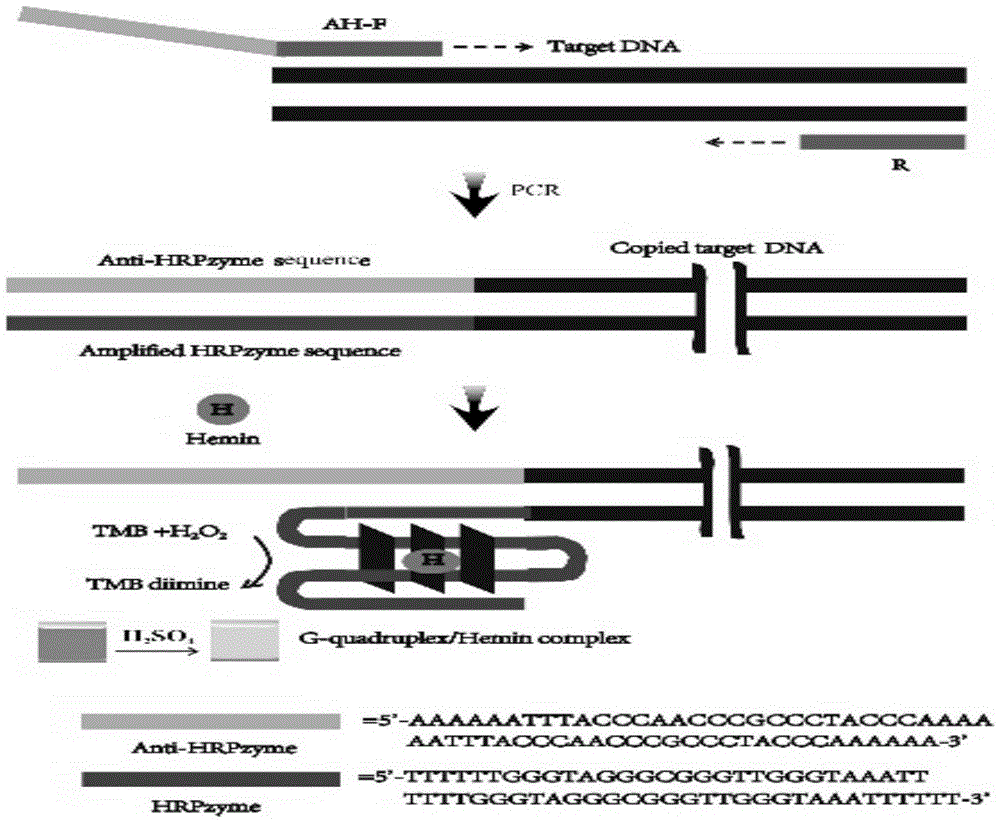
[0049] The difference is that the food pathogen to be tested is Campylobacter jejuni, the 5'end of the Campylobacter jejuni forward primer is linked to the horseradish peroxidase complementary sequence, and the gene sequence is AAAAAATTTACCCAACCCGCCCTACCCAAAAAATTTACCCAACCCGCCCTACCCAAAAA-5'- AGCAGGGATAAGCCCTCTTG-3'; the gene sequence of the reverse primer of Campylobacter jejuni is 5'-AGCGATCTATTTGCCATCG-3'.
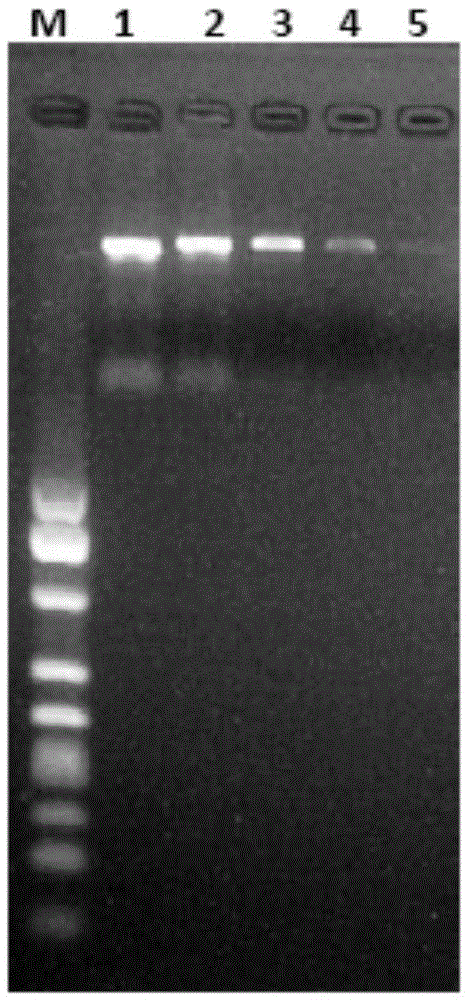
[0050] The DNA agarose gel electrophoresis diagram of the Campylobacter jejuni DNA template extracted by the magnetic bead method is as follows Picture 9 As shown in the figure, M, 1, 2, 3, 4, and 5 respectively represent DNA fragments with different molecular weights, and 1.3*10 is extracted 5 , 1.2*10 6 , 1.0*10 7 , 4.1*10 7 , 8.2*10 7 Electrophoretic band of cfu / mL Campylobacter jejuni DNA.
[0051] Picture 10 It is the agarose gel electrophoresis diagram of different concentrations of Campylobacter jejuni DNA template after PCR amplification reaction. M, 1, 2, 3, 4, 5 rep...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com