Construction and application of novel RNA cyclization expression vector
A technology of circularization and construction, applied in the biological field, can solve the problems of cumbersome construction process and limited types of circular RNA molecules
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0082] Example 1 Construction and Detection of pZW1-Circ-Vector Circular RNA Molecular Expression Vector
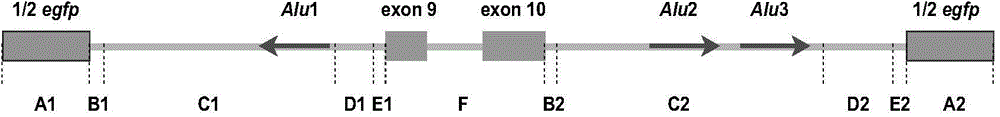
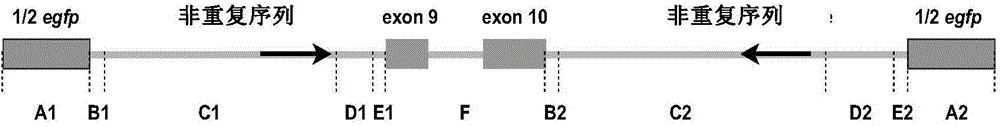
[0083] According to the current molecular structure and production mechanism of circular RNA molecules derived from exons, the present invention constructs a circular RNA molecule expression vector named pZW1-Circ-Vector, and uses Northern Blot and other experimental methods to verify the expression of circular RNA molecules. In this example, the 9th and 10th exons of the POLR2A gene are taken as examples to describe its construction method and verification implementation process.
[0084] 1. Experimental materials
[0085] 1.1 Experimental reagents
[0086] Restriction endonucleases Nhel and MluI were purchased from NEB Company in the United States; T4 DNA ligase and PrimeSTAR high-fidelity PCR system were purchased from Takara Company in Japan; Reagent was purchased from Invitrogen, USA; AmpliScribe TM T7 Transcription Kits were purchased from Epicentre Company of ...
Embodiment 2
[0151] Example 2 Construction and detection of pZW1-Circ-Vector-High high-efficiency circular RNA molecule expression vector
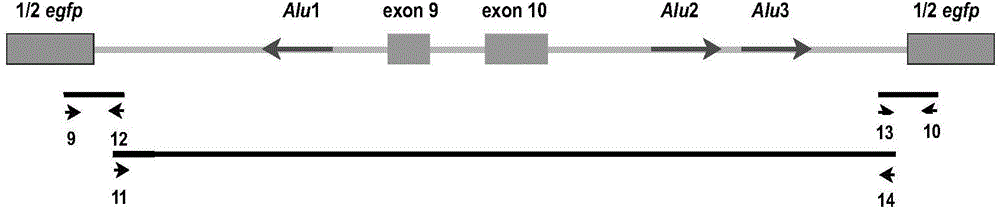
[0152] On the basis of the pZW1-Circ-Vector expression vector, the original Alu element can be replaced by two completely reverse complementary sequences, which can express exon-derived circular RNA more efficiently and obtain more circular RNA molecules. The optimized vector is named pZW1-Circ-Vector-High efficient circular RNA molecule expression vector. For a schematic diagram of plasmid construction see image 3 . The specific implementation process is as follows:
[0153] 1. Experimental materials
[0154] 1.1 Experimental reagents
[0155] With reagent used in embodiment 1.
[0156] 1.2 Cell lines and vectors
[0157] In this example, the H9 cell line of human embryonic stem cells and the HeLa cell line of human cervical cancer cells are still used. For details, please refer to 1.2 Cell lines and vectors in Example 1. The vector used to co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com