PCR detection primers for seven different species of cells and detection method and use thereof
A technology for detecting primers and cells, applied in the field of cell detection, can solve the problems of high cost, narrow species range, time-consuming and labor-intensive, etc., and achieve high sensitivity, strong specificity, and broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Example 1. Primers designed for PCR detection of cells (MDBK, MDCK, BHK21, ST, SP20, 293, VERO) derived from seven different species
[0051] Mitochondrial DNA (mtDNA) is often used as the target gene in PCR detection, and the inventors analyzed and confirmed that the cytochrome b (Cytb) gene in mitochondrial DNA is more suitable as the target gene for species identification. This is because the full length of the Cytb gene is about 1100-1200bp, and there is no significant difference in sequence length between species, but there is a bias in nucleotide composition and there are differences between species. The coding sequence region of Cytb gene evolves slowly, is relatively conserved, and exists in all mammals.
[0052] Based on the above ideas, the inventor retrieved the nucleotide sequences of the cytochrome b (Cytb) gene of seven different species of cells (MDBK, MDCK, BHK21, ST, SP20, 293, VERO) on Genbank, and used bioinformatics The method compares the nucleotid...
Embodiment 2
[0060] Embodiment 2, establish the PCR detection method of the cell (MDBK, MDCK, BHK21, ST, SP20, 293, VERO) of seven kinds of different species sources
[0061] The present invention uses the primers in Example 1 to carry out PCR detection on seven different species-derived cells (MDBK, MDCK, BHK21, ST, SP20, 293, VERO), including the following steps:
[0062] 1) Extract the genomic DNA of the cells to be tested
[0063] Seven kinds of cells, MDBK, MDCK, BHK21, ST, SP20, 293 and VERO, were selected from the laboratory to extract genomic DNA. The specific steps are as follows:
[0064] (1) Take two tubes of cryopreserved MDBK, MDCK, BHK21, ST, SP20, 293 and VERO 7 kinds of cells, thaw the cryopreserved tubes in warm water at 37°C, and collect the cells in 1.5mL centrifuge tubes, 8000rmp Centrifuge for 10min, discard the supernatant, and resuspend the cells with 1mL PBS;
[0065] (2) Take 200 μL of the resuspended cells into a new 1.5 mL centrifuge tube, add 1 mL of DNAzol Ly...
Embodiment 3
[0072] Embodiment 3, the specific detection of primer and detection method of the present invention
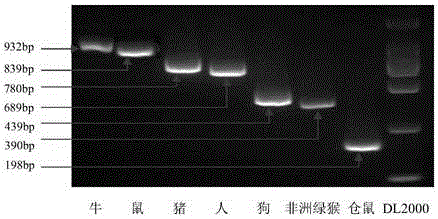
[0073] Using the genomic DNA of cells from seven different species (MDBK, MDCK, BHK21, ST, SP20, 293, VERO) at a concentration of 10 ng / μL as templates, the seven templates were tested with the seven pairs of primers designed in Example 1, respectively. PCR amplification to detect the specificity of each pair of primers. The PCR reaction system and PCR reaction procedure were the same as those in Example 2. After the PCR reaction was completed, the PCR products were detected by 1.5% agarose gel electrophoresis, and the loading volume was 10 μL.
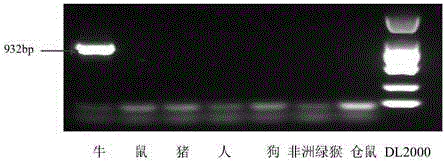
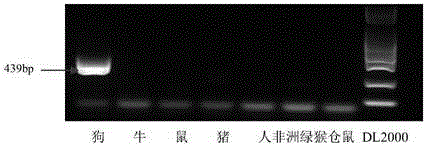
[0074] The result is as Figure 2-Figure 8 Shown:
[0075] figure 2 It is the specific detection result of the bovine-derived primer pair (C-F and C-R) on 7 kinds of cellular genomic DNA templates. The results show that the bovine-derived specific primers only amplify a 932bp fragment on the DNA template of bovine-derived MDBK cel...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com