Primer sequence and method for rapid identification of pseudosciaena crocea, larimichthys polyactis and collichthys lucidus
A primer sequence, technology of large yellow croaker, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problems of breeding, production and sales confusion, scarcity of wild large yellow croaker, and high price. The effect of shortening the technical cycle, the identification method is simple and fast, and the steps are simple
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] (1) Sample DNA was extracted and dissolved in double distilled water (100ng / ul), and stored at 4°C for later use.
[0027] (2) PCR amplification conditions
[0028] The numbers and sequences of the specific primers are:
[0029] Primer 1F sequence: 5'-GGAAAGAGCCAGGAAAGC;
[0030] Primer 1R sequence: 5'-GGCGGAACTCTGAGCAAA. (design according to EST sequence in GenBank)
[0031] The total volume of the PCR reaction is 25 μL, 10×Buffer 2.5 μl, dNTP 0.5 μl, Mg 2+ 1.5 μl, Taq enzyme 0.25 μl, each primer 1 μl (0.5 mmol / L), sample DNA template 1 μl, dH 2 O 17.25 μl.
[0032] The PCR amplification parameters were as follows: 94°C pre-denaturation for 5 minutes followed by 35 cycles: 94°C for 40 sec, 58°C for 40 sec, 72°C for 1 min; and finally 72°C for 10 min.
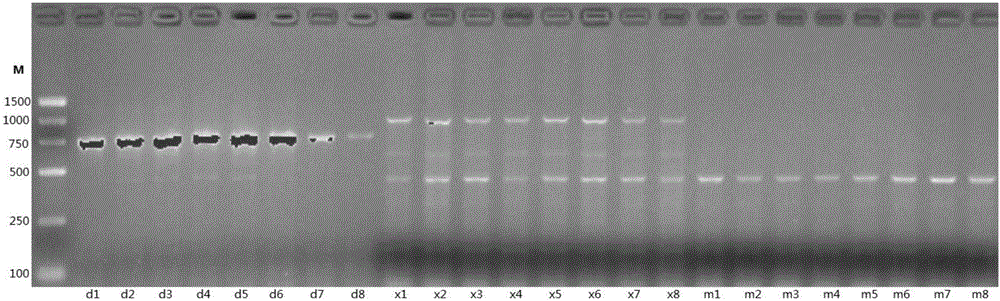
[0033] (3) Electrophoresis detection and map analysis of PCR amplification
[0034] Take 8 μL of PCR amplification products for detection on the electrophoresis instrument, the electrophoresis buffer is 0.5×TBE, t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com