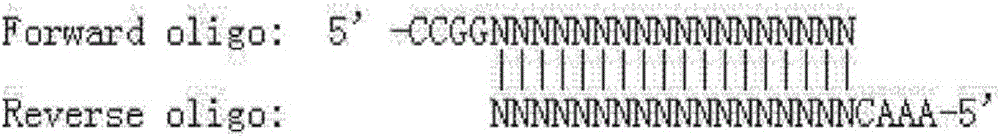CRISPR-Cas9 system and application therefore in treating breast cancer diseases
A breast cancer and multipurpose technology, applied in the field of genetic engineering, can solve the problems affecting the specific knockout of the target gene, wrong targeting, etc., and achieve the effect of wide applicability, low cost and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] The design of embodiment 1sgRNA
[0059] First, select the 5'-GGN(19)GG sequence based on the RecQL4 gene shown in SEQ ID NO: 1. If there is no 5'-GGN(19)GG sequence, 5'-GN(20)GG or 5'- N(21)GG also works. The targeting site of the sgRNA on the STAT6 gene is located in the exon of the gene. Use BLAT in the UCSC database or BLAST in the NCBI database to determine whether the target sequence of the sgRNA is unique. At the same time, according to the design rules of other sgRNAs, a total of 57 sgRNAs were designed. However, the final experiment confirmed that only 17 of them have the function of targeted modification. Here, the sequences without functions are not listed one by one, only given Two counter-examples, which also fully demonstrate that in the prior art, the design of sgRNA cannot obtain functional sgRNA only according to the design rules without experiments. The sgRNA sequence involved in the present invention is as follows:
[0060] RecQL4-sg1: gcaagcgcggag...
Embodiment 2
[0079] Embodiment 2, construct the oligonucleotide duplex of sgRNA
[0080] According to the selected sgRNA: RecQL4-sg1, add CCGG to its 5' to get the forward oligonucleotide
[0081] (Forward oligo) (if the sequence itself already has 1 or 2 Gs at the 5' end, then correspondingly omit 1 or 2 Gs); according to the selected sgRNA, obtain the complementary strand of its corresponding DNA, and at its 5' AAAC was added to obtain a reverse oligonucleotide (Reverse oligo). Synthesize the above-mentioned forward oligonucleotide and reverse oligonucleotide respectively, and the designed forward oligonucleotide (Forward oligo) is:
[0082] ccggcaagcgcggaggccgggcgggcg (SEQ ID NO: 21); the reverse oligonucleotide (Reverse oligo) is aaacgcccgcccggcctccgcgcttgc (SEQ ID NO: 22).
[0083] The forward oligo and reverse oligo of the synthesized sgRNA oligonucleotide are denatured and annealed in pairs, and after annealing, a double strand that can be connected to the U6 eukaryotic expression...
Embodiment 3
[0086] Embodiment 3, the construction of sgRNA oligonucleotide plasmid
[0087] 1. Linearize the pGL3-U6-sgRNA plasmid. Enzyme digestion system and conditions are as follows: 2 μg pGL3-U6-sgRNA (400ng / μl); 1 μl CutSmart Buffer; 1 μl BsaI (NEB, R0535L); replenish water to 50 μl, incubate at 37 degrees for 3-4 hours, shake and centrifuge at intervals To prevent droplets from evaporating onto the tube cap.
[0088] 2. Ligate the annealed sgRNA oligonucleotide double strands to the linearized pGL3-U6-sgRNA plasmid to obtain the pGL3-U6-RecQL4-sg1 plasmid.
[0089] 3. Transform and coat Amp+ plates (50 micrograms / ml).
[0090] 4. identify positive clones with the method of universal primer U6 sequencing, the primers are
[0091] atggactatcatatgcttaccgta.
[0092] 5. Shake the bacteria overnight on a 37-degree shaker and extract the pGL3-U6-RecQL4-sg1 plasmid with a plasmid extraction kit.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com