A detection method and device for low-frequency gene fusion
A detection device and gene fusion technology, which are applied in the fields of biochemical cleaning devices, biochemical equipment and methods, and microbial determination/inspection, etc., and can solve the problems of short length of truncated sequences, wrong distinction, difficult library construction or sequencing, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
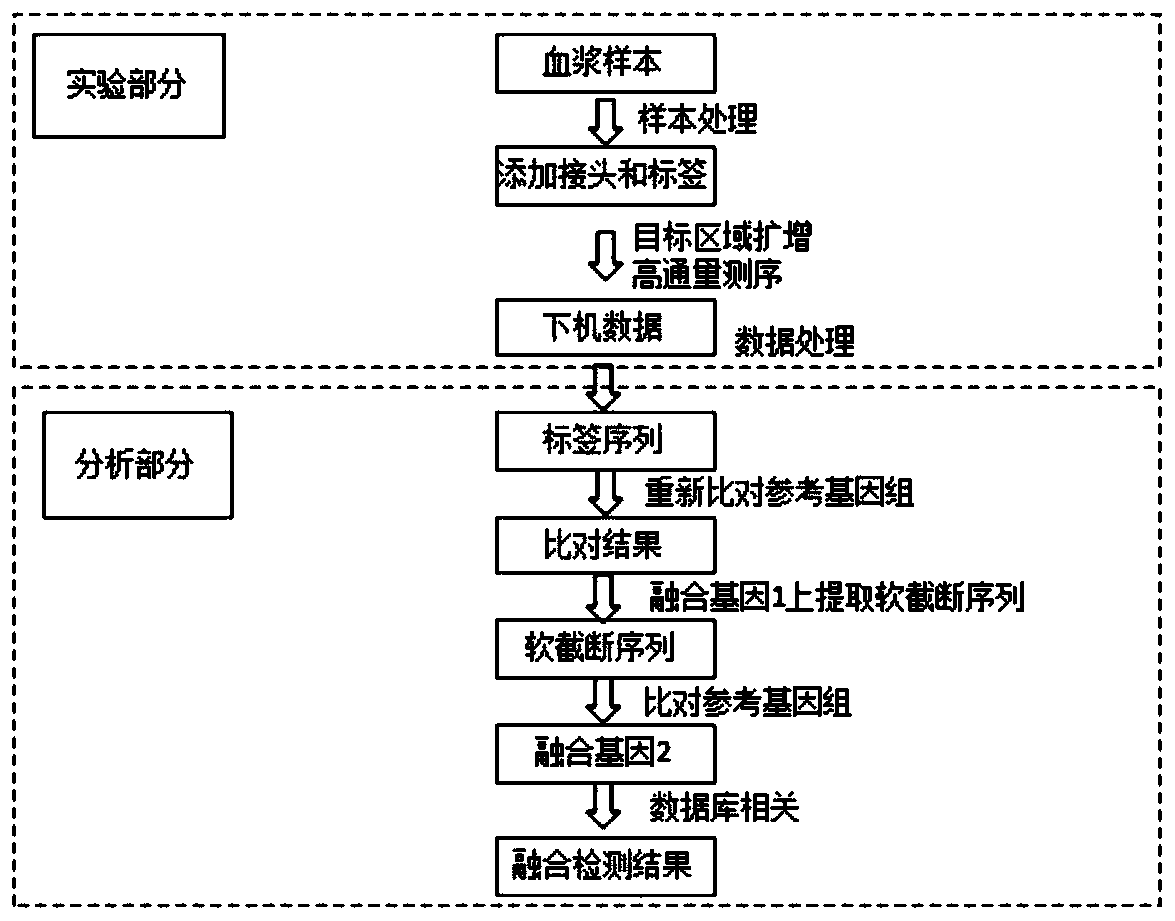
[0026] According to a typical implementation of the present invention, S8 includes: S81, use bwa software to compare the data after getting off the machine to the reference genome using bwa software, and use samtools software to sort and build an index; S82, compare the files Perform traversal, merge the two sequences with the same label at each site into one, and remove any inconsistent sequence compared with most other sequences in the same label; Back to the reference genome, and use samools software for sequencing and indexing. Synthesizing sequences with the same label can restore the sequence to its original condition, eliminate errors in PCR, and aligning and indexing can speed up subsequent processes.
[0027] According to a typical embodiment of the present invention, the variation detection analysis in S9 includes: S91, synthesize a long soft truncated sequence from all soft truncated soft truncated positions close to each other obtained in S83, and use the sequence ...
Embodiment 1
[0037] The samples used in this example are plasma samples from patients with non-small cell lung cancer, which are positive plasma samples for gene fusion, and the tissue RNA of the plasma samples detects EML4-ALK.E6bA20.AB374362 fusion by PCR.
[0038] Detect according to the method of the present invention below.
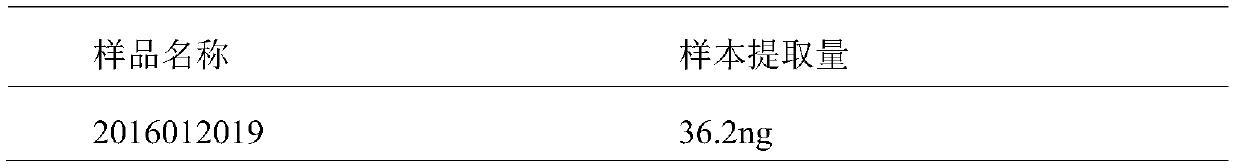
[0039] 1. 10ml of whole blood from patients with non-small cell lung cancer is collected and transported in BCT tubes from Streck Company. The transport temperature is room temperature and the transport time does not exceed 72 hours. Plasma was separated by two-step centrifugation, that is, centrifuged at 1600g for 10 minutes, the supernatant was taken, and then centrifuged at 16000g for 10 minutes. The supernatant was the separated plasma, which was stored at -80°C. The cfDNA in plasma was extracted using Qiagen’s circulating DNA (circulating) extraction kit, and the extracted cfDNA was stored at -20°C for later use. See Table 1 for sample names and extraction ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com