Molecular markers related to ear length traits of millet and their detection primers and applications
A millet and ear length technology, applied in the field of molecular markers and their detection primers and applications, can solve the problems that have not been reported
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1: High-throughput sequencing and SNP marker information analysis
[0030] In this example, the materials of the male parent "Zhang Zagu No. 3" and the female parent "A2", the F1 generation materials obtained by crossing the two, and the 441 materials of the F2 population obtained after 13 generations of self-crossing of the F1 generation, that is, recombinant self-crossing Department (Recombinant inbred lines, RILs). Each material was planted in Zhangjiakou, Hebei, and the trait data such as ear length were recorded and sorted. After the degenerate genome resequencing of each material, the alignment of the reference genome was completed, and the SNP marker was obtained. By analyzing the data of ear length traits, the associated SNP markers were obtained, and the SNP markers were verified by clonal sequencing.
[0031] In this example, the RADseq method was used to extract the genomic DNA of each sample individual (including 441 RILs, 2 parents, and 1 F1 indiv...
Embodiment 2
[0061] Example 2: SNP marker verification
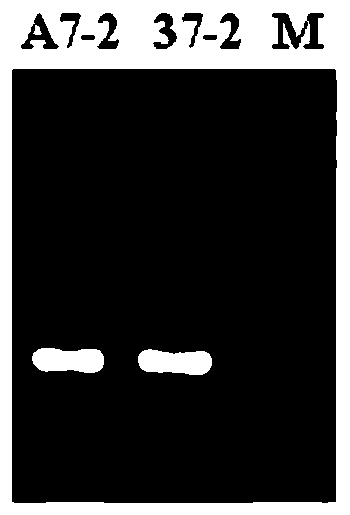
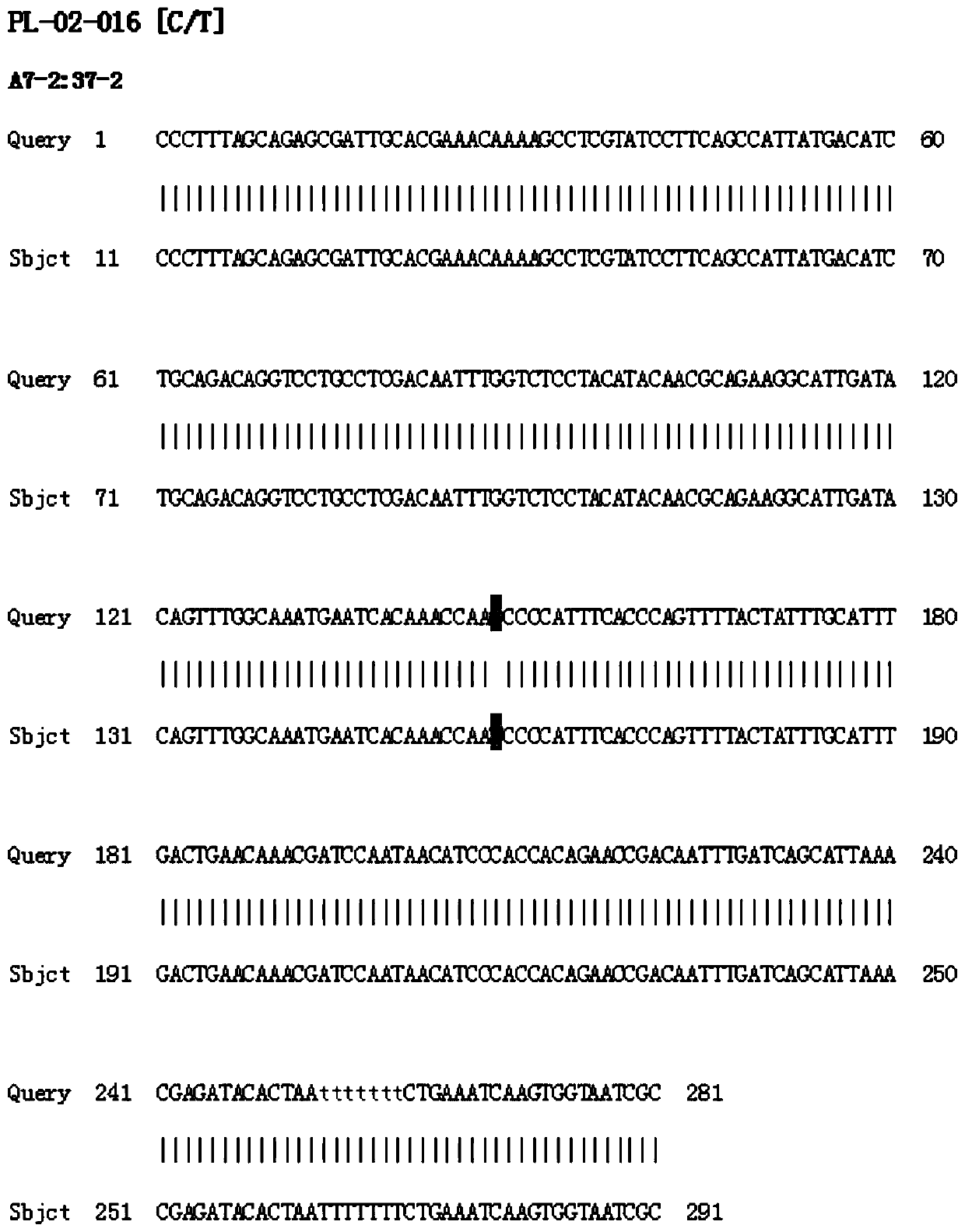
[0062] According to the predicted bin interval and SNP site, compare the reference genome to obtain the gene sequence of the relevant region, select about 300 bp before and after the SNP site, design and develop SNP marker primers, and use the DNA of the paternal and maternal materials as templates for PCR amplification. Select primers that amplify normally and PCR products conform to the predicted size of the amplification product, recover the product and perform sequencing, and select labeled primers with SNP site differences in the amplified gene sequences of the paternal and maternal materials.
[0063] The predicted bin markers contain multiple SNP sites, and these sites are screened according to the PCR results.
[0064] First, according to the sequencing results after PCR product recovery, the markers with SNP site differences in the amplified gene sequences of the paternal and maternal materials were selected. Specific steps...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com