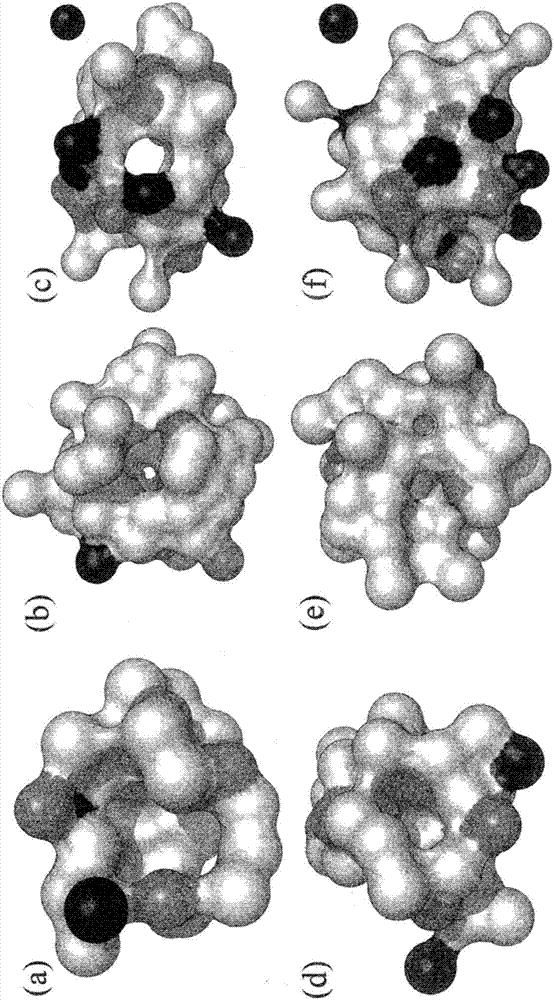
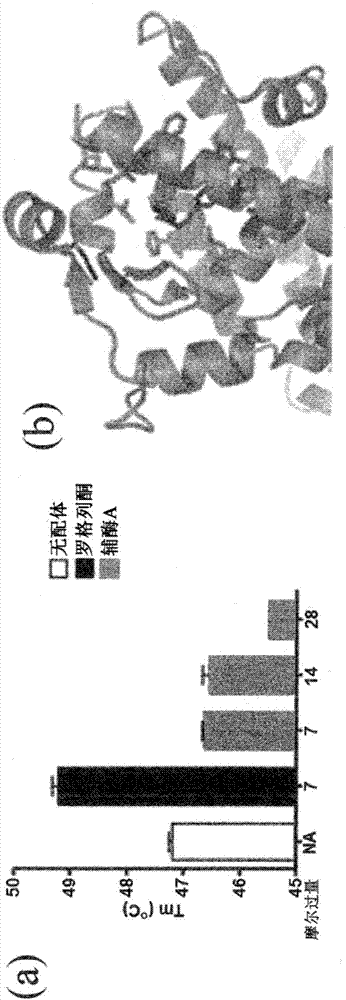
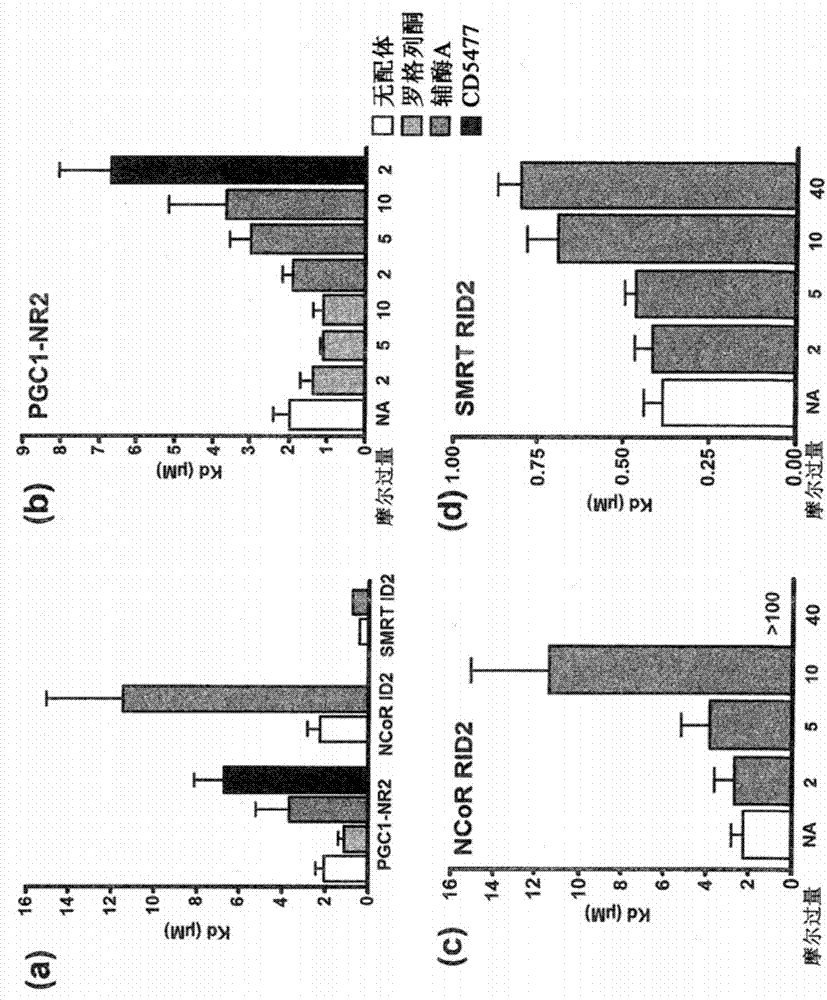
Methods and systems for identifying ligand-protein binding sites
A technology for binding sites and proteins, which is applied in biochemical equipment and methods, determination/inspection of microorganisms, and analysis of two-dimensional or three-dimensional molecular structures. Sexuality, not predicting known drug targets, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0132] An Integrated Structure and Systems Based Framework for the Identification of Metabolites and New Targets of Known Drugs
[0133] Build a Probabilistic Pocket Ensemble (PPE)
[0134] Drugs that are inherently promiscuous may bind to different protein pockets with various characteristics, making it difficult to establish a general description of where a drug might bind. To capture the essential binding site features of promiscuous drugs, we established a method for constructing PPE of such drugs (see Methods section for details). The PPE represents a unified collection of individual pockets that may bind to several conformations of the drug. Each position in the PPE can consist of multiple atoms from different residues. The frequency of atoms and residues at each position is recorded and used to construct the maximum probable sequence similarity scoring function. This probability scoring method properly takes into account the fact that a drug can bind to several pocke...
example 2
[0266] Evaluation of PPE and Probability Scoring Function
[0267] To investigate the ability of PPE to retrieve structurally similar drug-binding pockets, we compared the protein pocket bound by 2′-adenosine monophosphate 5′-ribose diphosphate to the predicted binding pockets of the top 10 predicted targets for this compound. Our sequence order-independent alignment overlaps with these 10 predicted targets, and the average normalized RMSD of the drug PPE for this construct is (Alignments contain >4 atoms). In contrast, using sequence order-relative structure alignment, only 2 predicted pockets (within PDB id 2BXP and 3ELM) coincided with the established drug pocket, and the average normalized RMSD was Likewise, using sequence order-relative alignments, the top 10 targets align on average to 9.5 atoms establishing the drug pocket, with an average RMSD of In contrast, with PPE, the average alignment length is 12.4 atoms, and the average RMSD is This result suggests that ...
example 3
[0285] new drug discovery
[0286] For new drugs, our method can be used following the protocol described below.
[0287] 1. Potential binding partners can be identified from a list of commonly observed protein targets (eg, Nature Reviews Drug Discovery 5, 821-834 (October 2006)).
[0288] 2. To test the binding of soluble drugs to recombination to obtain sufficient quantities of the initial list of proteins, we can utilize isothermal titration calorimetry experiments or surface plasmon resonance. For proteins that are not readily available, or drugs that require special solvents (such as DMSO), we can use microthermophoresis or differential scanning fluorescence. Many known protein drug targets are readily available commercially, and many expression plasmids are available in the nonprofit ADDGENE database.
[0289] 3. In order to obtain the binding site of a new drug to one of the above proteins (with sufficiently high interaction strength), we can perform x-ray crystallogr...
PUM
| Property | Measurement | Unit |
|---|---|---|
| affinity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com