Drug relocation method based on gene mutation and gene expression
A gene expression and gene expression profiling technology, applied in the field of data mining, can solve the problem of low accuracy of positioning results, and achieve the effect of improving the accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0034] The present invention will be described in further detail below in conjunction with the accompanying drawings and specific embodiments.
[0035] This embodiment takes liver cancer as an example.
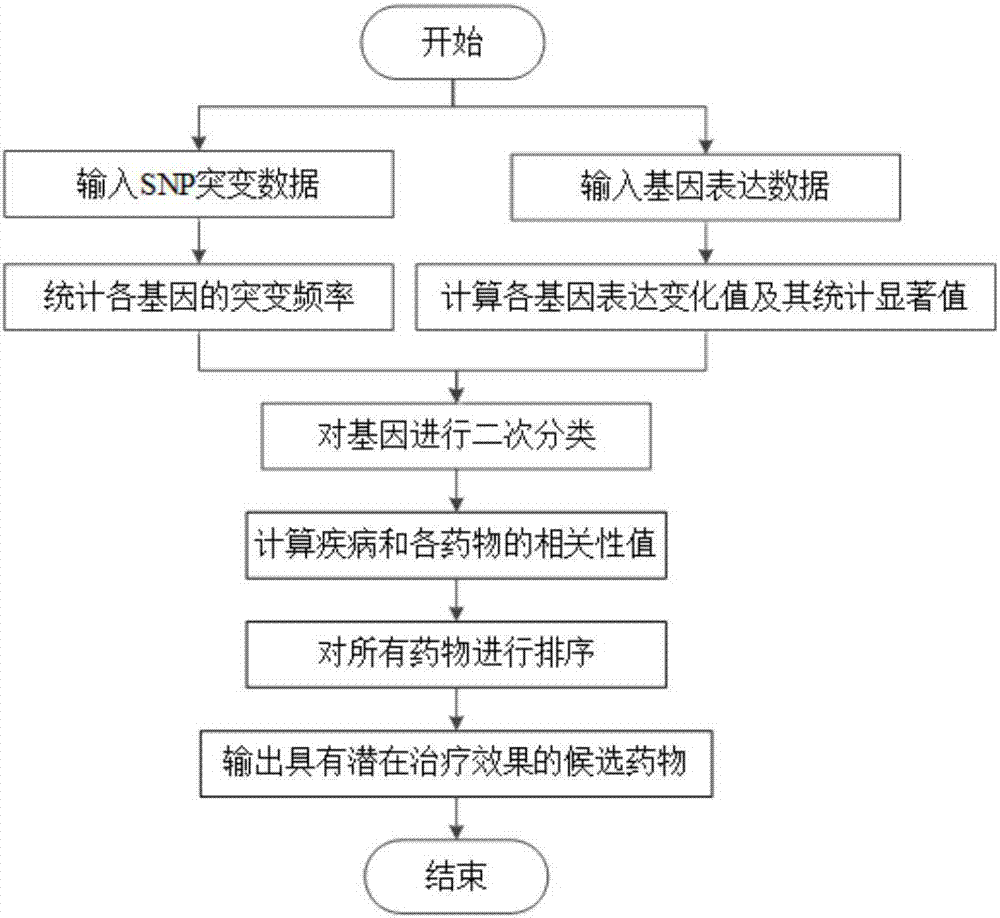
[0036] refer to figure 1 . A drug repositioning method based on gene mutation and gene expression, comprising the following steps:
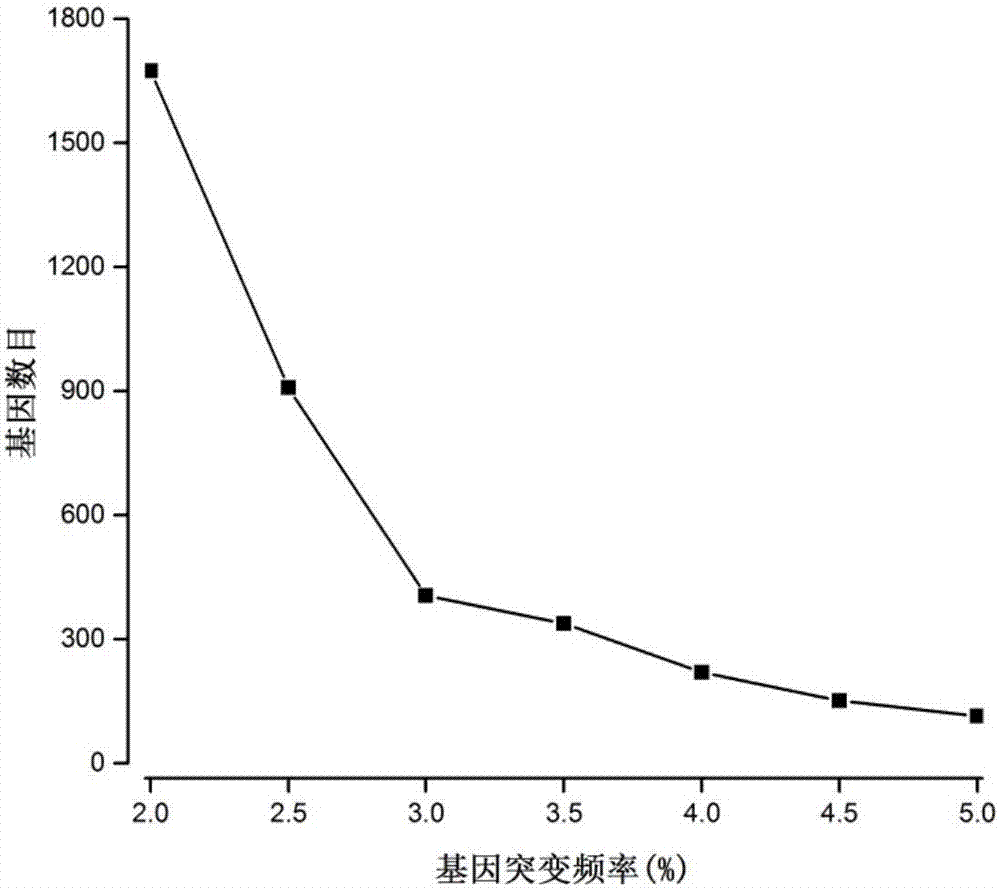
[0037] Step 1) processing SNP mutation data: count the mutation frequency of each gene in all disease state samples in the SNP mutation data, and obtain the mutation frequency data of all genes;
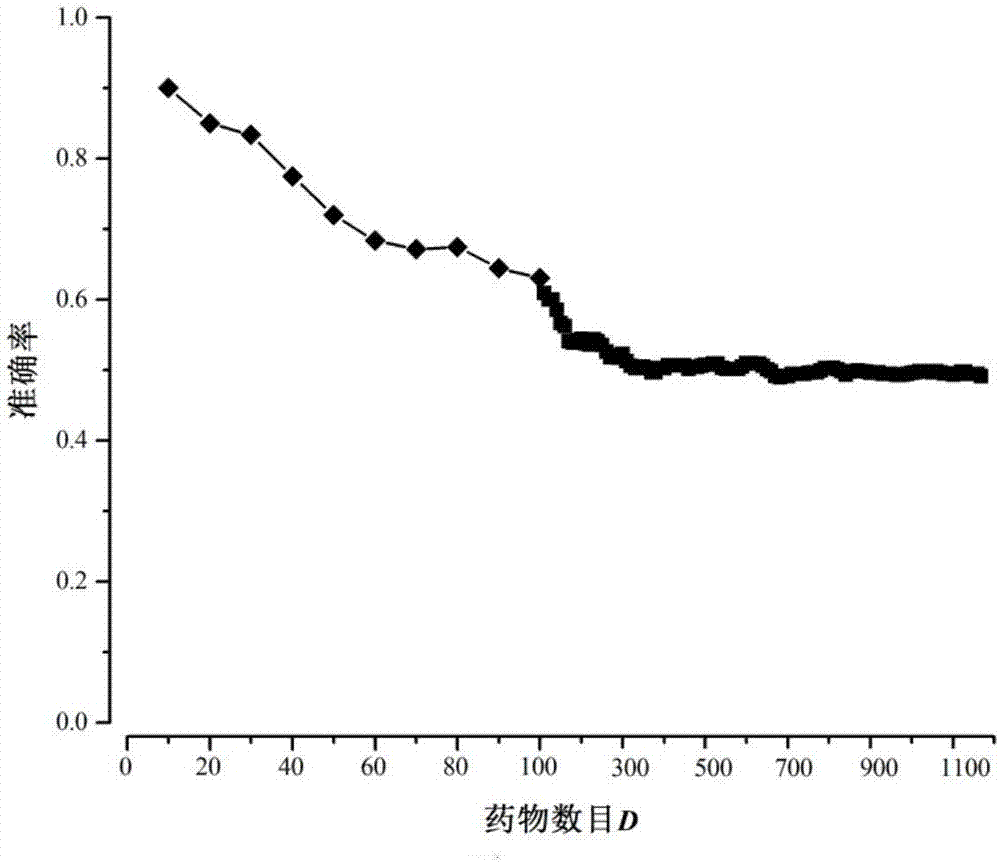
[0038] Step 2) Process the gene expression data, and calculate the change value of each gene expression and its statistical significance:
[0039] (2a) Obtain the cancer patient sample set and the normal person sample set: use the Barcode code provided by TCGA to identify the sample type of the gene expression data in TCGA, specifically: the digital code of the fourth part of the Barcode code of the sample is from 01 to 09 The interval is a cancer pa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com