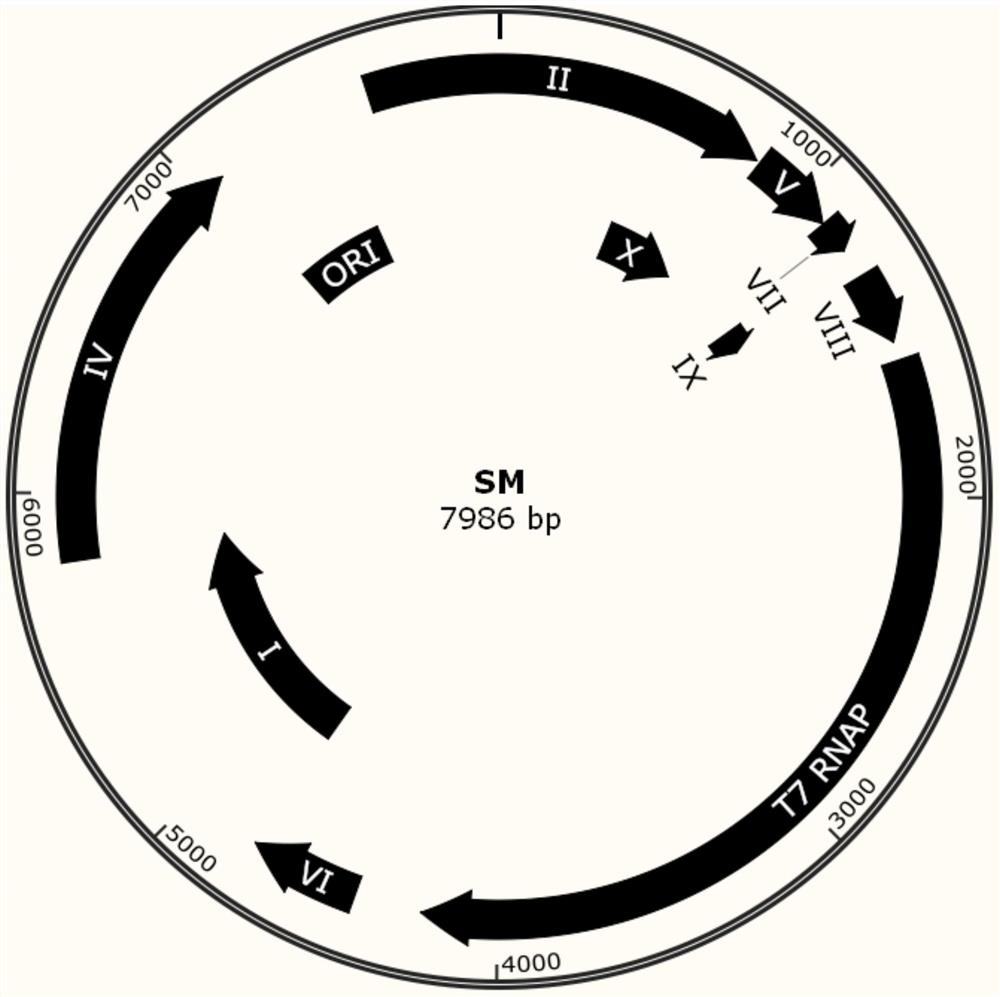
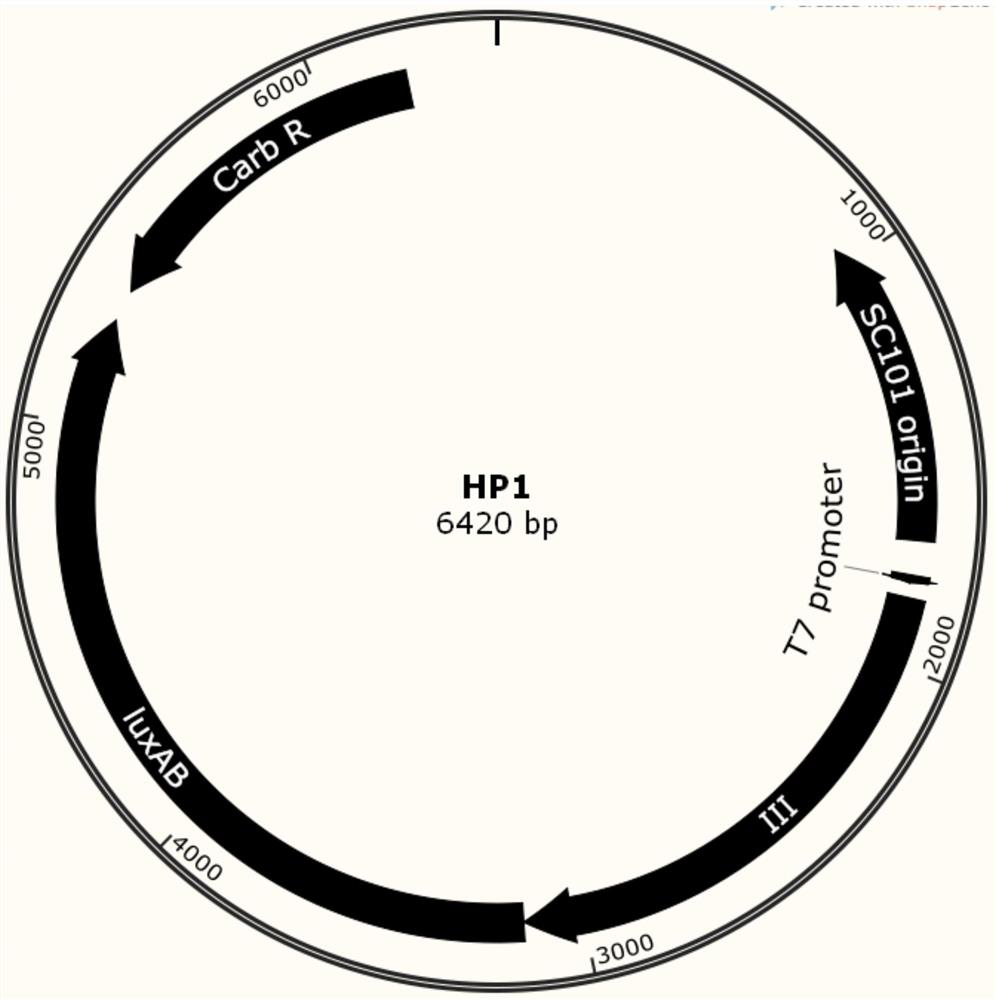
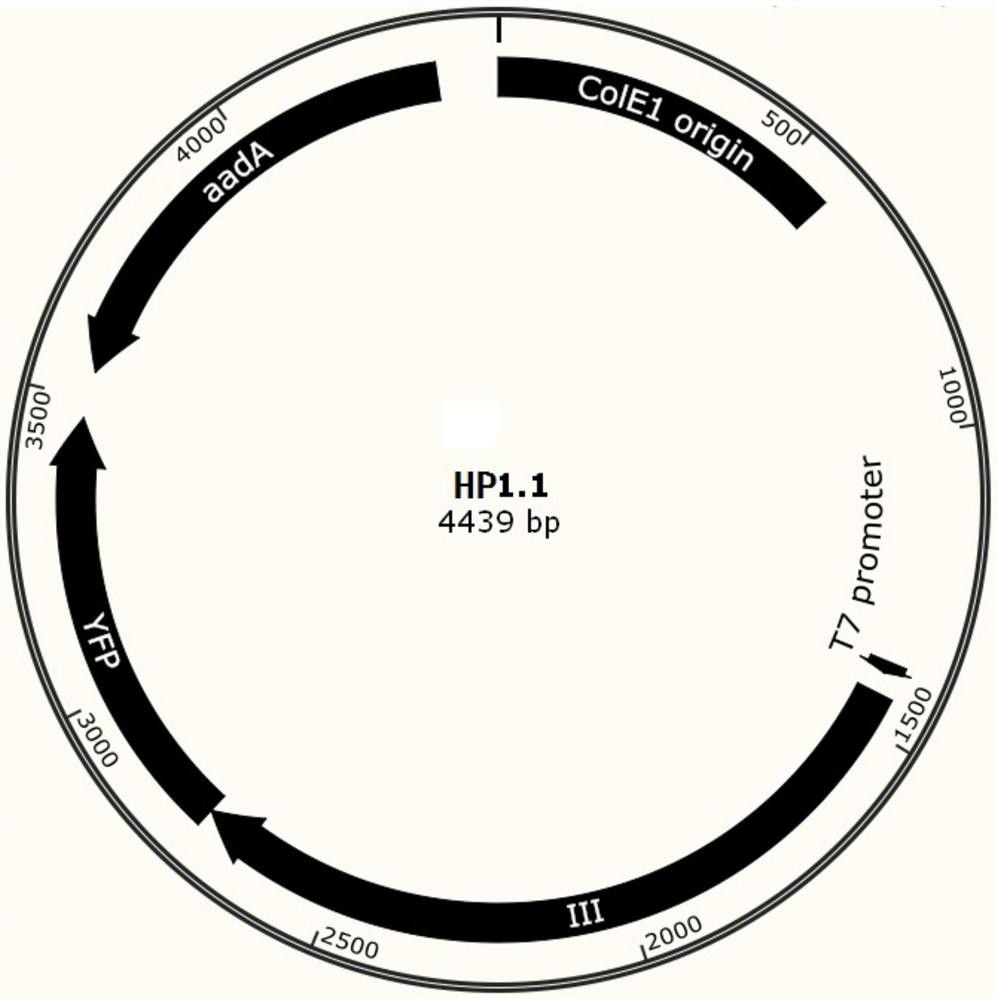
A phage-assisted continuous directed evolution system and method for multiple bacteria
A directed evolution and phage technology, applied in biochemical equipment and methods, viruses/phages, bacteria, etc., can solve the problems of mutual interference of different genetic elements and low efficiency of phage proliferation and evolution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068]Example 1 Evolution desired host fungi preparation
[0069]1) The host fungi S1 carrying HP1 and IP plasmids, in LB medium containing 50 μg / ml of carboxylcin and 25 μg / ml chloramphenicol-resistant, to OD600 = 0.6 at 37 ° C, 220 rpm. The bacterial liquid is placed at 4 ° C for spare, and the preservation time is no more than 1 day.
[0070]2) The host bacteria S2 carrying HP2 and IP plasmids was cultured to OD600 = 0.6 at 37 ° C, 220 rpm in LB medium containing 50 μg / ml of carboenomycin and 25 μg / ml chloramphenicol. The bacterial liquid is placed at 4 ° C for spare, and the preservation time is no more than 1 day.
[0071]3) The host fungi S3 carrying HP2, HP3, and IP plasmids, in LB medium containing 50 μg / ml of carboenomycin, 50 μg / ml of magnifier and 25 μg / ml chloramphenicol resistance, at 37 ° C, 220 rpm Conditions were cultured to OD600 = 0.6.12000 rpm 1min centrifugal collected bacteria, and then resuspended in an equal volume of 50 μg / ml of carboenomycin and 25 μg...
Embodiment 2
[0077]Example 2S5 SMP observation method
[0078]1) In a 10 cm bacterial culture plate, a layer of 10 ml of 2% agarose gel was paved, and 20 min was placed at room temperature to solidify.
[0079]2) When the SM concentration is unknown, it is necessary to press SM to press 0 times and 10.1,2,3,4,5The proportion of times is diluted in a continuous gradient.
[0080]3) Take 6 groups of 200 μl of Example 1, S5 host bacteria prepared, each group added to SM 10 μl of different dilution gradients in "2), and then 4 mL preserved at 55 ° C containing 0.4% agar lb medium and final The concentration of 50 μg / ml carboxylmycin. After mixing the vortex shock mixer, the sample is placed into the plate prepared in "1)".
[0081]4) Culture overnight in a biochemical incubator at 37 ° C.
[0082]5) Observe the number of plazots formed by the statistical gradient dilution sample, and the SM concentration is calculated.
Embodiment 3
[0083]Example 3S6 and SM plaque observation method in S7 host bacteria
[0084]Similarly to Example 2, the S6 bacterium formed in Example 1 was replaced with the S6 bacterium, which was prepared in Example 1, which was replaced with the second embodiment.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com