Multi-target sequence sgRNA expression vector based on endogenous tRNA processing system and application of sgRNA expression vector in plant gene editing
A technology for expressing vector and RNA polymerase, applied in the direction of DNA/RNA fragment, introduction of foreign genetic material and vector using vector, etc., which can solve the problems of dicotyledonous plant report and increase of cost, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
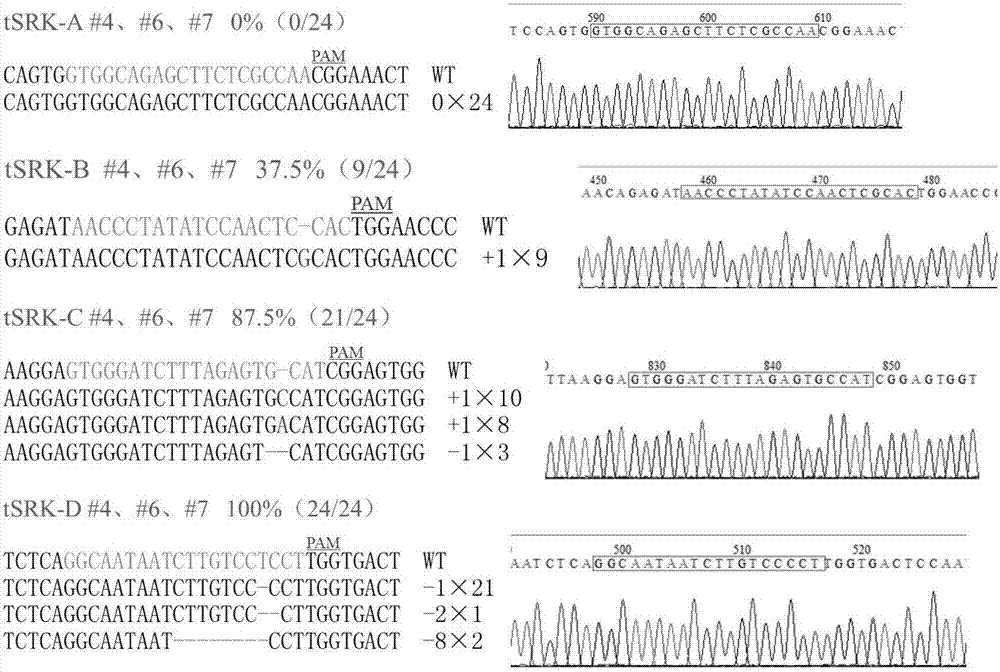
[0056] Example 1 Using tRNA-sgRNA sequence expression to realize mutation of cabbage self-incompatibility related gene SRK
[0057] 1. Artificially synthesize the U6-26::tRNA-sgRNA expression cassette sequence shown in the present invention.
[0058] 2. According to the SRK3 haplotype gene sequence of the cabbage self-incompatibility line F416 and the target site sequence requirements of the CRISPR / Cas9 gene editing system, four target sites of the SRK3 gene A, B, C, and D were selected and designed. The following target site complementary primers anneal to double-stranded DNA fragments.
[0059] (1) SRK3 gene target site A complementary primer sequence:
[0060] sgSRK-1F:5-TGCAGTGGCAGAGCTTCTCGCCAA-3
[0061] sgSRK-1R:5-AAACTTGGCGAGAAGCTCTGCCAC-3
[0062] (2) SRK3 gene target site B complementary primer sequence:
[0063] sgSRK-2F:5-TGCAAACCCTATATCCAACTCCAC-3
[0064] sgSRK-2R:5-AAACGTGGAGTTGGATATAGGGTT-3
[0065] (3) SRK3 gene target site C complementary primer sequence...
Embodiment 2
[0076] Example 2 Using tRNA-sgRNA sequence expression to realize the mutation of tobacco PDS gene
[0077] 1. Artificially synthesize the U6-26::tRNA-sgRNA expression cassette sequence shown in the present invention.
[0078] 2. According to the PDS gene sequence of common tobacco and the target site sequence requirements of CRISPR / Cas9 gene editing, three target sites of PDS gene A, B, and C were selected, and the complementary primers for the target sites were designed as follows to anneal to double-stranded DNA fragments .
[0079] (1) NtPDS gene target site A complementary primer sequence:
[0080] sgNtPDS1F:5-TGCAGCCGTTAATTTGAGAGTCCA-3
[0081] sgNtPDS1R:5-AAACCGGACTCTCAAATTAACGGC-3
[0082] (2) NtPDS gene target site B complementary primer sequence:
[0083] sgNtPDS2F:5-TGCAGCTGCATGGAAAGATGATGA-3
[0084] sgNtPDS2R:5-AAACTCATCATCTTTCCATGCAGC-3
[0085] (3) NtPDS gene target site C complementary primer sequence:
[0086] sgNtPDS3F:5-TGCAGAGGCAAGAGATGTCCTAGG-3
[0...
Embodiment 3
[0095] Example 3 Using tRNA-sgRNA sequence expression to realize the double mutation of cabbage MS1 gene and SRK gene
[0096] 1. Artificially synthesize the U6-26::tRNA-sgRNA expression cassette sequence shown in the present invention.
[0097] 2. According to the MS1 gene (male sterility-related gene) sequence of the cabbage self-incompatibility line F416 and the sequence requirements of the target site of the CRISPR / Cas9 gene editing system, four of the MS1 genes, A, B, C, and D, were selected. For the target target site, design the following target site complementary primers to anneal to double-stranded DNA fragments.
[0098] (1) MS1 gene target site A complementary primer sequence:
[0099] sgMS1-1F: 5-TGCAGCCTTTCTAAAACTAGAAGG-3
[0100] sgMS1-1R: 5-AAACCCTTTCTAGTTTTAGAAAGGC-3
[0101] (2) MS1 gene target site B complementary primer sequence:
[0102] sgMS1-2F: 5-TGCAGAGTAGCAAAAGGAGAGCCG-3
[0103] sgMS1-2R: 5-AAACCGGCTCTCCTTTTGCTACTC-3
[0104] (3) MS1 gene target...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com