Deep learning based long-chain non-coding RNA subcellular position prediction algorithm
A long-chain non-coding, deep learning technology, applied in the field of long-chain non-coding RNA subcellular location prediction algorithms, can solve the problem of bias and poor performance in the main category, and achieve the effect of avoiding over-fitting problems.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0069] The following is a detailed description of the embodiments of the present invention. This embodiment is carried out based on the technical solution of the present invention, and provides detailed implementation methods and specific operation processes.
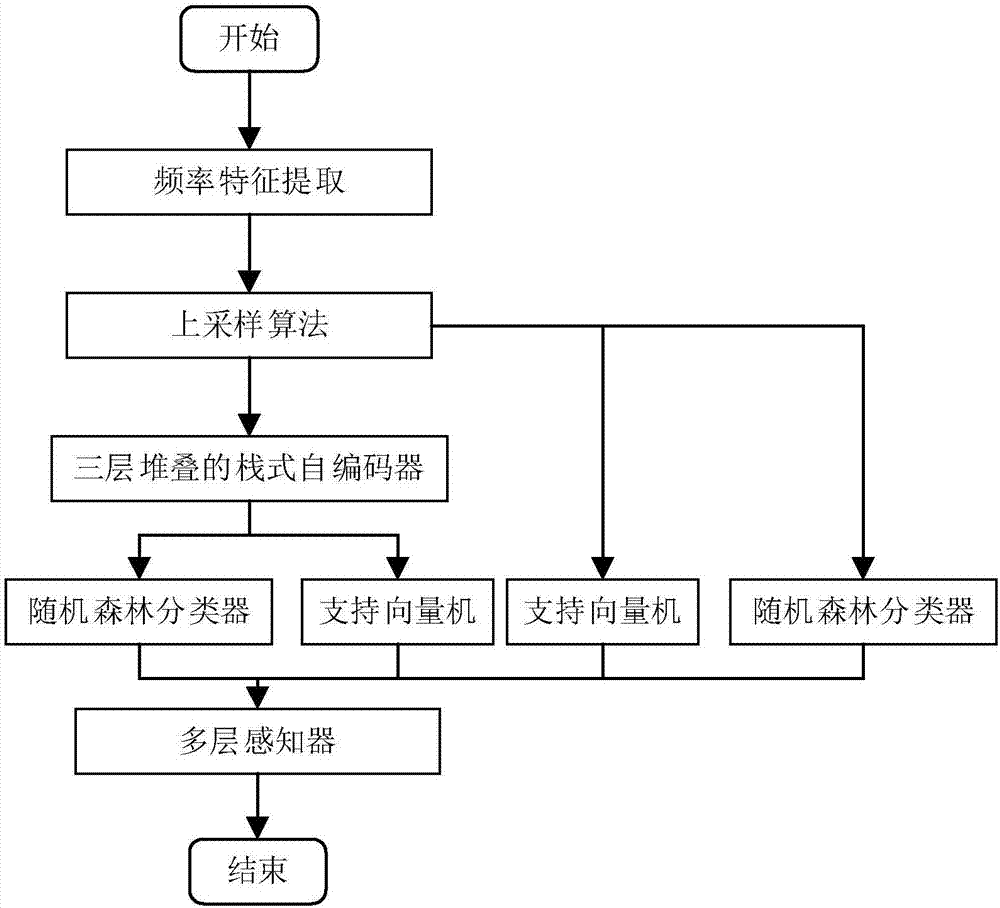
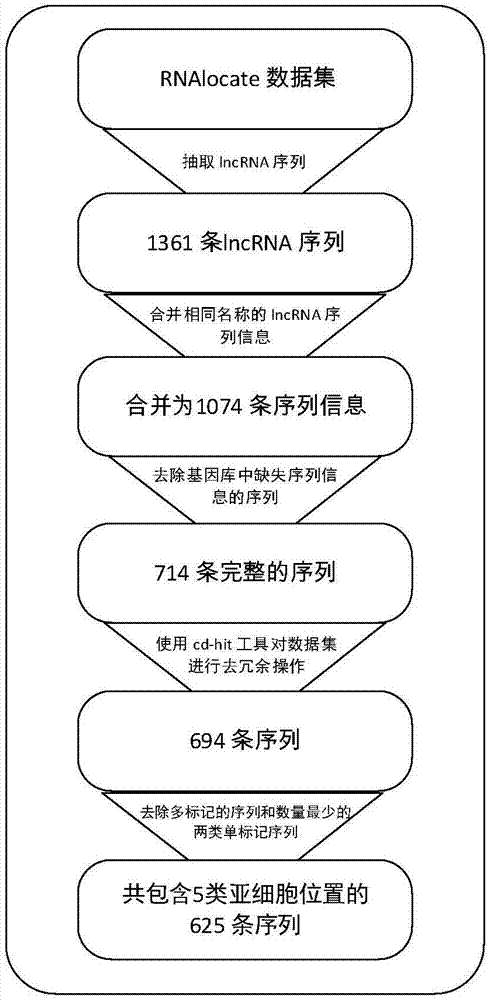
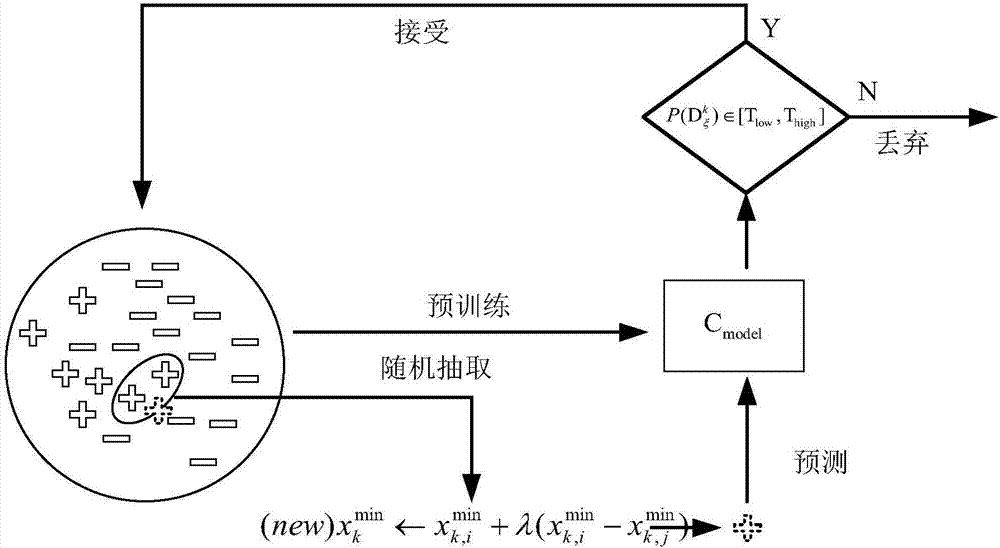
[0070] The present invention takes into account the imbalance of the data set, wherein the number of samples located in the cytoplasm, nucleus, cytosol, ribosomes and exosomes is 304, 152, 96, 47 and 26 respectively, so for each category except the first category double upsampling. The activation functions adopted by the encoding layer and the decoding layer in the three-layer stacked self-encoder in the present invention are all sigmoid functions, the adam optimizer selected by the optimizer, and the square error between the reconstructed output and the original input selected by the loss function. Both Batch_size and nb_epoch are 100, and the number of neurons in the three layers is set to 256, 128, and 64 respectivel...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com