Method and primer for detecting mutation of ORF15 exon of RPGR gene
A technology of exons and sequencing primers, applied in the direction of DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of low accuracy of sequencing results, mixed peaks of sequencing results, and difficult sequences, so as to enhance integrity and accuracy, The effect of high sensitivity and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] The inventor designed a large number of primers for exon ORF15 of the RPGR gene, and screened primers with good specificity through optimization and comparison of primer reaction conditions. The primers are shown in Table 1.
[0038] Table 1 The present invention provides amplification primers and sequencing primer sequences and their positions on the chromosome
[0039]
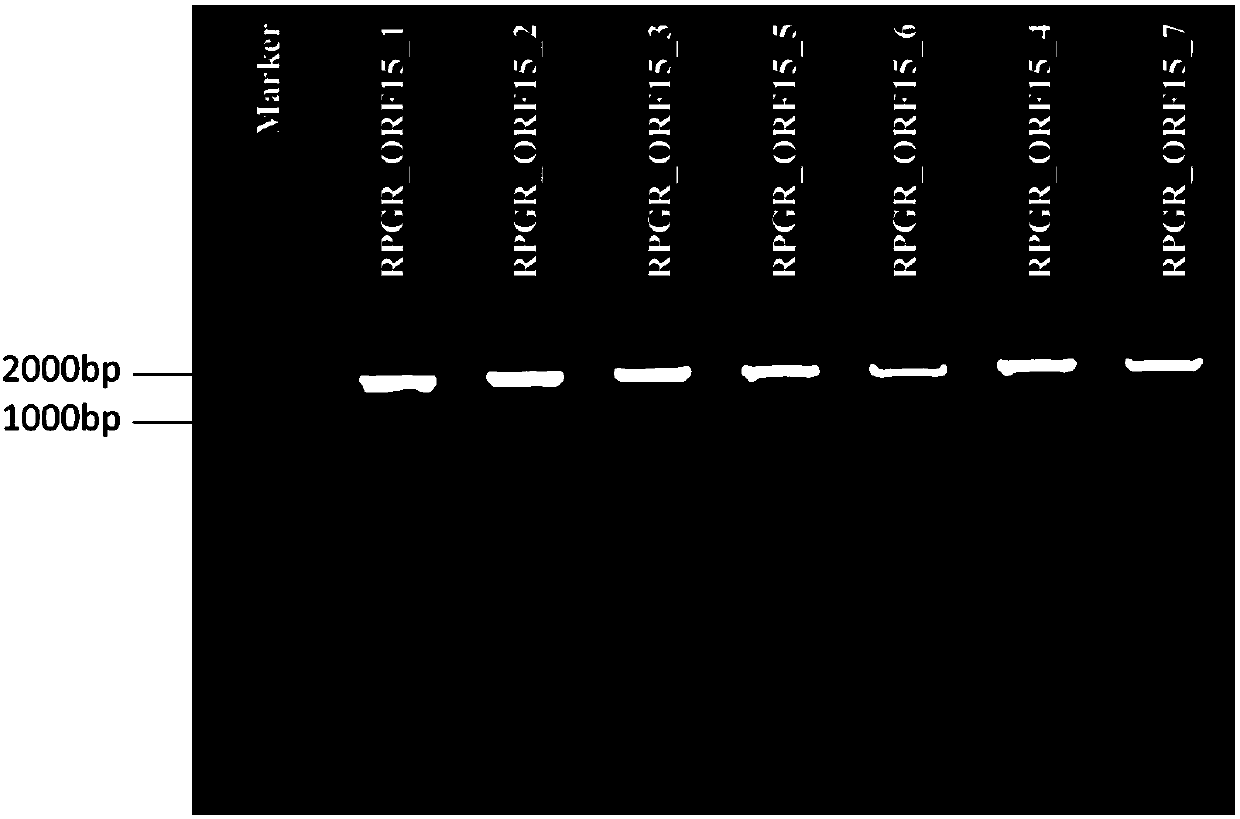
[0040] The seven sequencing primers of the present invention were blasted in UCSC, and the amplified fragment of the RPGR gene ORF15 exon primer was located between chrX: 38144608-38146560, with a length of 1953 bp. No other homologous genes, the results are as follows figure 1 It is consistent with the reference sequence of RPGR gene.
[0041] The detection method of RPGR gene ORF15 exon mutation comprises the following steps
[0042] (1) Genomic DNA was extracted using a blood DNA extraction kit (Biteco, DP1801) according to the instructions of the kit.
[0043] The brief operation of extracti...
Embodiment 2
[0071] Example 2 Detection of RPGR gene ORF15 exon mutations in clinical samples
[0072] Three clinical family samples were operated as described in Example 3, DNA extraction, amplification, electrophoresis detection, product purification, sequencing reaction, and result analysis were performed to obtain sequencing results. Such as Figure 10 As shown, the patient has RPGR:c.3031G>T:(p.G1011X) mutation, the patient's father is normal at this site, and the patient's mother is a carrier.
Embodiment 3
[0073] Example 3 compares the accuracy of high-throughput sequencing
[0074] (1) According to the operation described in Example 3, operations such as DNA extraction, amplification and electrophoresis detection were performed.
[0075] (2) Purification of PCR products: DNA purification kit (QIAGEN, Cat. No. 28004) was used to purify PCR products according to the operating instructions. The brief steps are as follows:
[0076] 1) Add 250 μl of Buffer PB to 50 μl of PCR product and mix well.
[0077] 2) Add the mixed solution to the MinElute elution column, centrifuge at 17900×g (13000 rpm) for 1 min at room temperature, and discard the effluent.
[0078] 3) Add 750 μl of Buffer PE to the MinElute elution column, centrifuge at 17900×g (13000 rpm) for 1 min at room temperature, and discard the effluent.
[0079] 4) Centrifuge the MinElute elution column again at 17900×g (13000 rpm) at room temperature for 1 min.
[0080] 5) Put the MinElute elution column into a new centrifug...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com