Method for easily and rapidly evaluating efficiency of molecular cloning system
A molecular cloning and rapid technology, applied in the field of molecular biology, can solve the problems of invisible molecular experiments and low efficiency of vector construction, and achieve the effect of low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
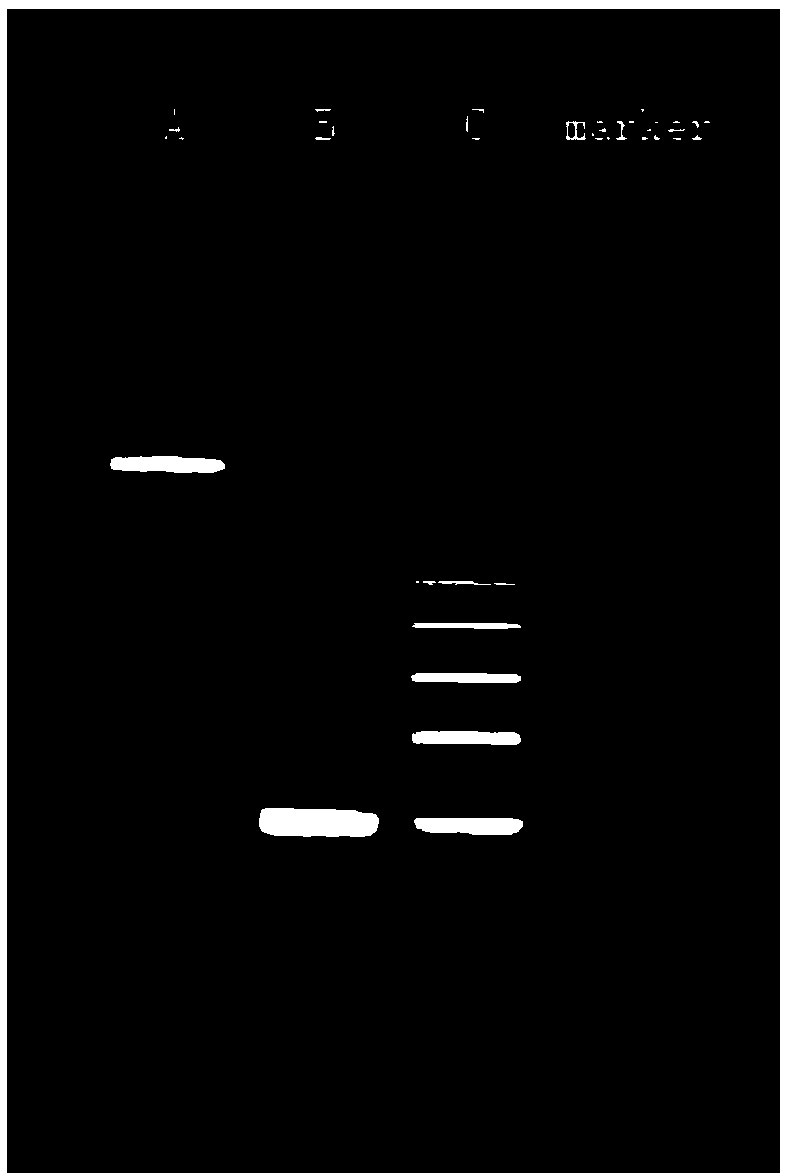
Examples
Embodiment 1
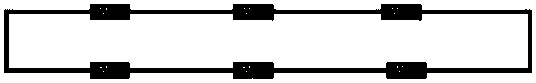
[0018] In this embodiment, a circular DNA fragment is designed, on which 6 multi-cloning sites MCS are arranged, and the sequence of one MCS is shown in SEQ ID NO.1 in the sequence listing; between multiple said multi-cloning sites MCS The intervals are equal in distance, denoted as K, specifically as figure 1 The pBRTEST DNA vector shown is obtained by entrusting a gene synthesis company with whole gene synthesis, as shown in SEQ ID NO.2 in the sequence listing.
[0019] Wherein, each multi-cloning site MCS has an enzyme cutting site for a restriction endonuclease to be tested, and the enzyme cutting site only exists on the multi-cloning site MCS; The sticky ends generated in the multiple cloning site MCS can be connected complementary, so that any two sticky ends can be connected to each other. The corresponding enzyme cleavage sites in MCS are EcoRI-SacI-KpnI-SmaI-BamHI-XbaI-SalI-PstI-SphI-HindIII. This vector can be single-digested with any of the above enzyme cleavage si...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com