PCR efficient amplification method of long-sequence gene without base mutation
A long-sequence, abasic technology, applied in the field of PCR amplification and TA cloning of long-sequence genes, can solve the problems of affecting protein function, low mismatch rate, missense mutation, etc., and improve the effect of obtaining accurate gene fragments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0024] The present invention is described in detail below in conjunction with embodiment. In order to make the object, technical solution and advantages of the present invention clearer and clearer, the present invention will be described in further detail below with reference to the accompanying drawings and examples, but the present invention is not limited to these examples. Unless otherwise specified, the raw materials and catalysts in the examples of the present invention were purchased through commercial channels.
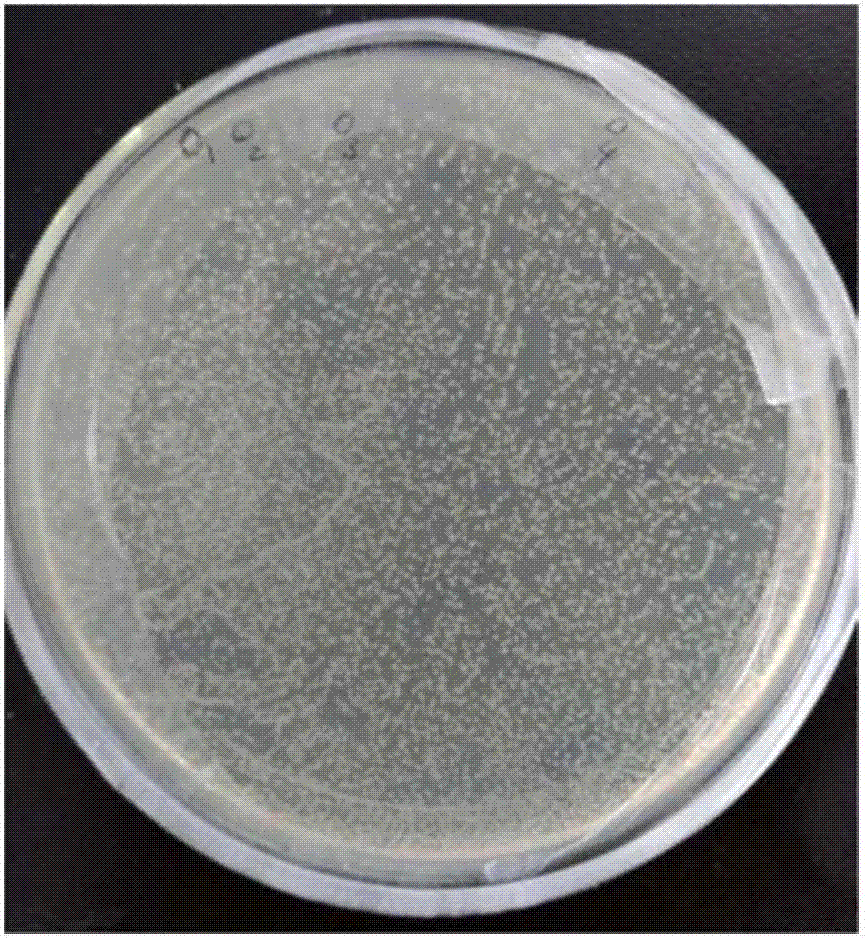
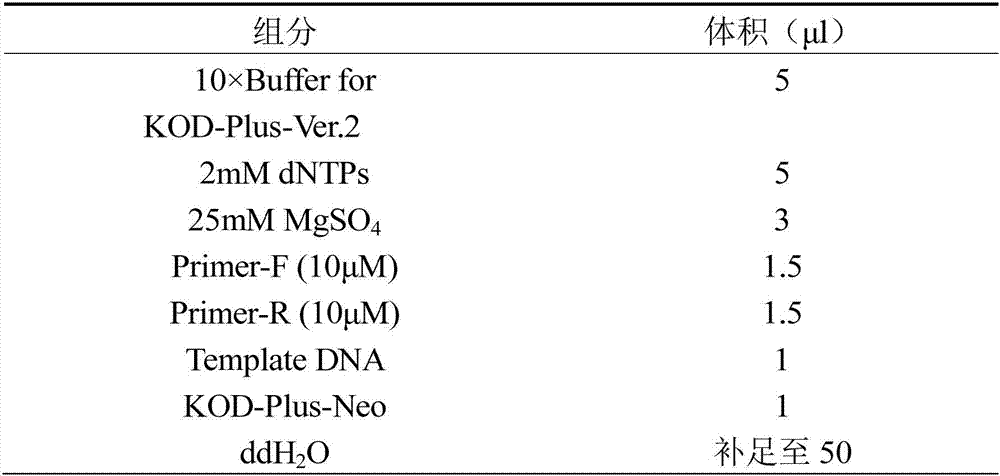
[0025] In the present invention, we amplify the complete gene sequence of rice EAT1, including the promoter and terminator of the EAT1 gene, all exons and introns of the EAT1 gene, and the length of the complete EAT1 gene expression cassette is 5225bp. According to the analysis of the base structure of the EAT1 gene, the GC% of the expression cassette of this gene is about 40%, and there is no problem of high GC. In the embodiment of the present invention, w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com