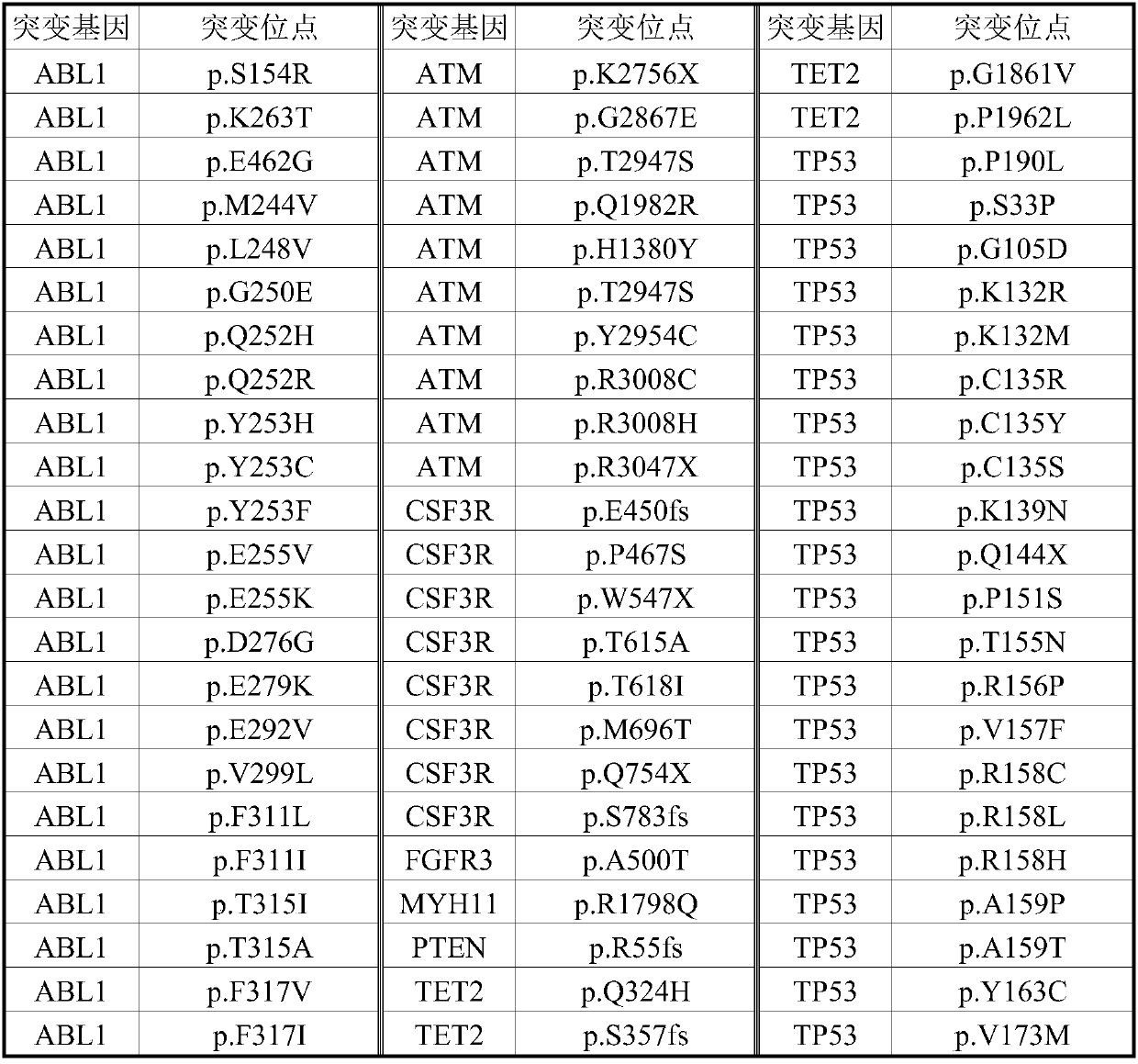
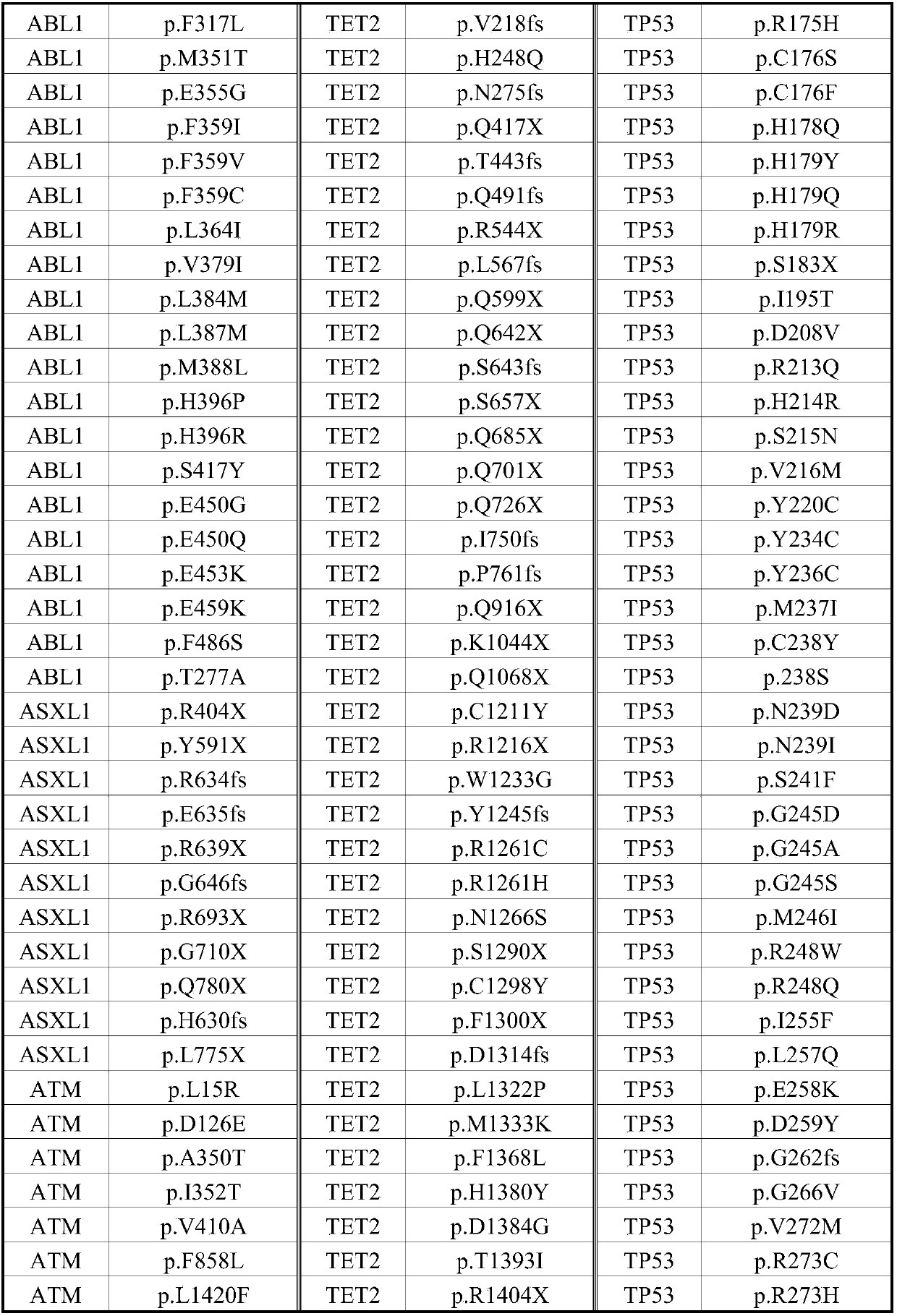
Detection kit for detecting related genetic group of CML
A detection kit and the technology of the kit, which are applied in the field of molecular biology, can solve the problems such as the limitation of the diagnosis method of chronic myeloid leukemia and the lack of diagnosis of chronic myeloid leukemia, and achieve the effect of high detection rate and high detection efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0031] Specific embodiments of the present invention will be described in detail below. In order to avoid too many unnecessary details, well-known structures or functions will not be described in detail in the following embodiments. Unless defined otherwise, technical and scientific terms used in the following examples have the same meaning as commonly understood by one of ordinary skill in the art to which this invention belongs.
[0032] Materials and methods:
[0033] Among the patients diagnosed from August 2015 to April 2017 at the Hematology Hospital of Chinese Academy of Medical Sciences and Union Medical Diagnostic Center, according to the following criteria, we conducted continuous enrollment (40 patients).
[0034] The inclusion criteria were: patients diagnosed with chronic myelogenous leukemia (CML) by cytomorphology and flow cytometry.
[0035] The diagnostic results of the 40 patients are shown in Table 3:
[0036] Table 3 List of patient diagnosis results
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com