Specific primer and probe as well as real-time fluorescence quantification PCR kit used for detecting DNA of peanut
A real-time fluorescence quantitative and specific technology, applied in DNA/RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc., can solve problems such as false positives, increased fluorescence intensity, and poor specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] The preparation of embodiment 1 Arah1 gene standard product,
[0036]To establish a real-time fluorescent quantitative PCR method, the external standard required by the method must first be prepared. The standard should contain highly conserved and specific sequences, and high specificity of the reaction must be ensured. The present invention uses peanut Arah1 rDNA as the target sequence. This example mainly uses PCR technology to amplify the Arah1 rDNA gene of peanut seeds, and uses gene recombination technology to connect it to the plasmid vector pMD18-T to construct the recombinant plasmid pMD18-T-Arah1, and carry out corresponding PCR identification and sequencing identification, Finally, it is quantified as a standard for the method to be established, laying the foundation for the next method and evaluation.
[0037] 1. Preparation of template DNA
[0038] 1. Genomic DNA was extracted from peanut seeds and used as a template for PCR amplification of the Arah1 rDN...
Embodiment 2
[0087] Embodiment 2 real-time fluorescent quantitative PCR kit
[0088] 1. Design and synthesis of specific primers and probes
[0089] A set of real-time fluorescent quantitative PCR primers and probes were designed using the Primer Premier5 software with the conserved fragment of Arah1 rDNA gene selected above as the target.
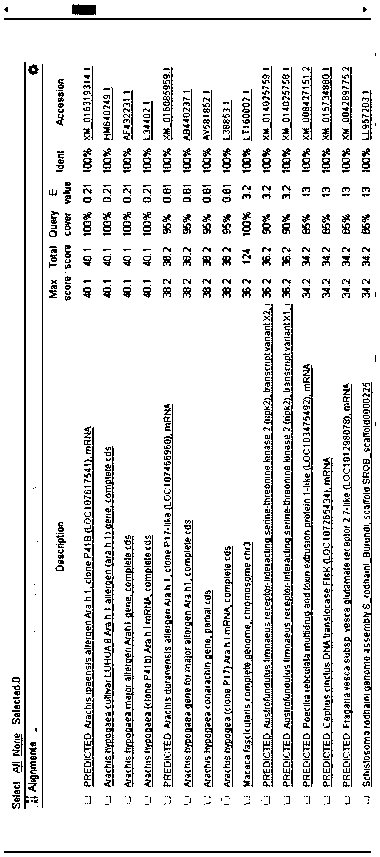
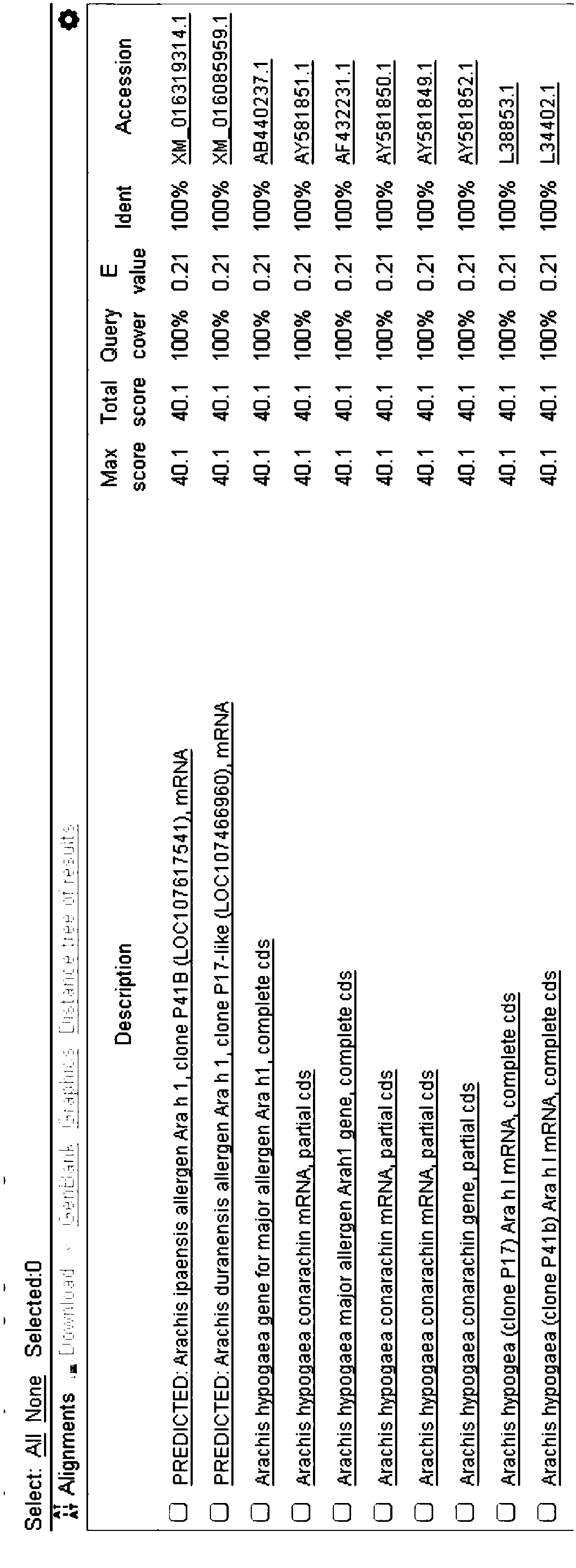
[0090] Arah1 gene of peanut oil was downloaded from NCBI, and multiple pairs of Arah1 gene primers and probes were designed by Primer Premier 5 and Becon Designer software, and primer specificity was verified by BLAST in NCBI. see the result Figure 1-Figure 3 .
[0091] Figure 1-Figure 3 It is the comparison result of 3 pairs of primer probe specificity BLAST.
[0092] Each pair of primers for peanuts is screened for primer specificity by ordinary PCR. The selected templates are: peanuts, soybeans, sunflowers, flax, rapeseed, corn, sesame, olives, walnuts, potatoes, negative for water, amplified bands Run the gel, and one pair of primers has the h...
Embodiment 3
[0137] Example 3 The quantitative detection method of the sample to be tested by the real-time fluorescent quantitative PCR kit
[0138] Respectively with the Arah1 rDNA gene series concentration standard substance (serial concentration standard substance series concentration standard substance) prepared by the sample DNA to be tested and embodiment 1: 1.00 * 10 8 copies / ml, 1.00×10 7 copies / ml, 1.00×10 6 copies / ml, 1.00×10 5 copies / ml,, 1.00×10 4 copies / ml) as a template, use the primers and probes in the kit to perform fluorescent quantitative PCR amplification, and set up positive and negative controls at the same time.
[0139] The PCR reaction system is as follows:
[0140]
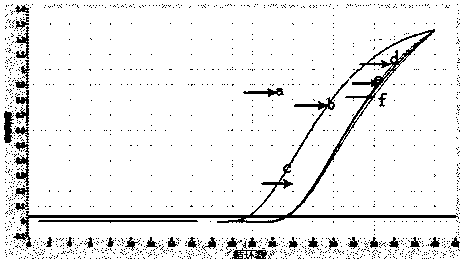
[0141] Reaction conditions: 94°C pre-denaturation for 2 minutes, 94°C for 15 seconds, 60°C for 40s and collecting fluorescence signals, 40 cycles. Draw a standard curve, and perform rapid quantitative detection through the standard curve and the Ct value of the sample to be tested.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com