Gene classification method and device
A classification method and gene technology, applied in special data processing applications, instruments, electronic digital data processing, etc., can solve the problem of low clustering effect, and achieve the effect of good clustering effect, strong generalization ability, and strong learning ability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
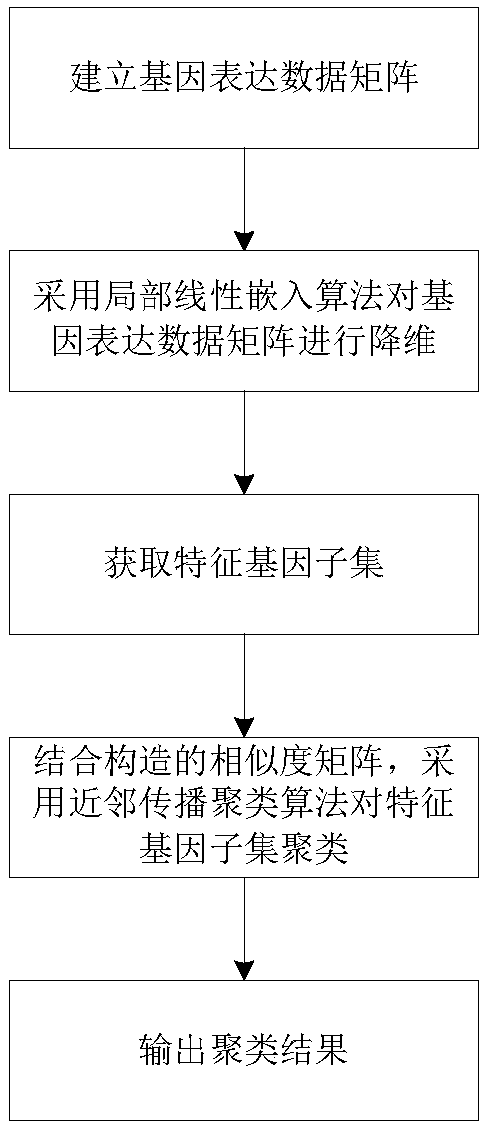
[0112] Such as figure 1 Shown, a kind of gene classification method of the present invention comprises the following steps:
[0113] Acquire gene expression data, the number of samples contained in the gene expression data is the first set value, the number of genes in each sample is the second set value, and the genes in the gene expression data are arranged and combined to form a matrix, the formed matrix is the gene expression data matrix.
[0114] Using a local linear embedding algorithm to reduce the dimension of the gene expression data matrix, calculate the linear embedding matrix of the gene expression data matrix, and obtain the feature gene subset after dimension reduction. That is, calculate the k nearest neighbors of all samples in the gene expression data matrix, construct a local reconstruction weight matrix according to the k nearest neighbors of each sample, and then use the local reconstruction weight matrix to calculate the gene expression data matrix Linea...
Embodiment 2
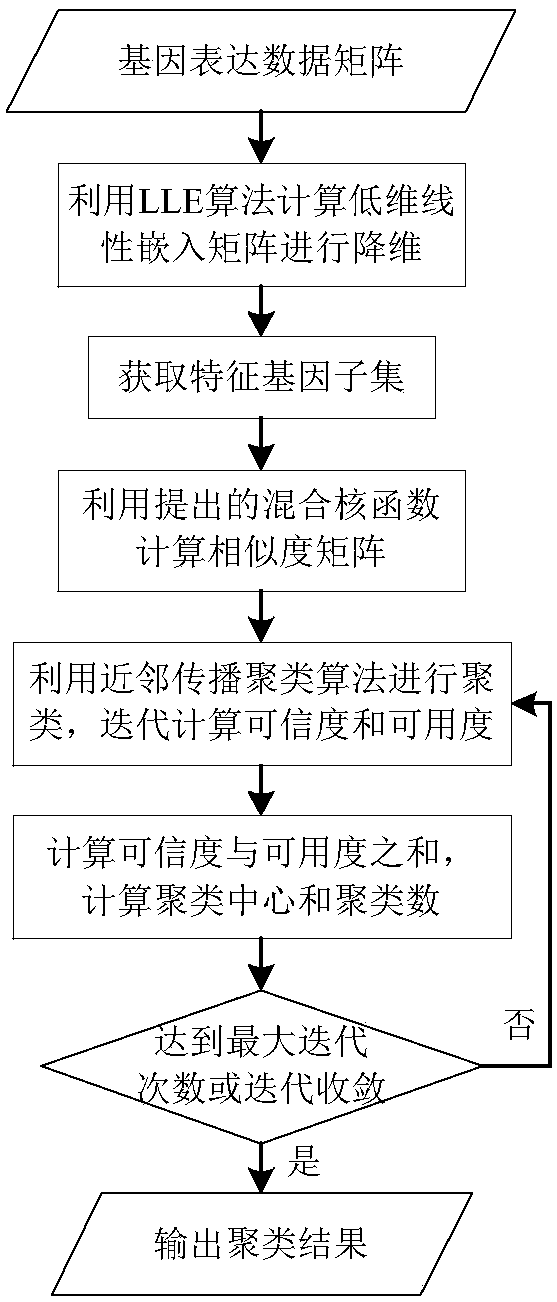
[0155] In order to avoid directly using the AP clustering algorithm to cluster the gene expression data set to obtain a large number of clusters, the present invention combines the LLE algorithm with the AP clustering algorithm based on the hybrid kernel function. First, the original high The three-dimensional gene data set is mapped to a low-dimensional space, and the characteristic gene subsets are obtained through linear dimension reduction; then the characteristic gene subsets after dimensionality reduction are clustered using the AP clustering algorithm based on the hybrid kernel function, and the final clustering is obtained result.
[0156] Such as figure 2 As shown, the specific steps are as follows:
[0157] Data preprocessing: use the genetic data acquisition system to obtain the original gene expression data set, including the gene expression values of multiple samples and the gene expression data matrix of the sample class label. The description of the gene exp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com