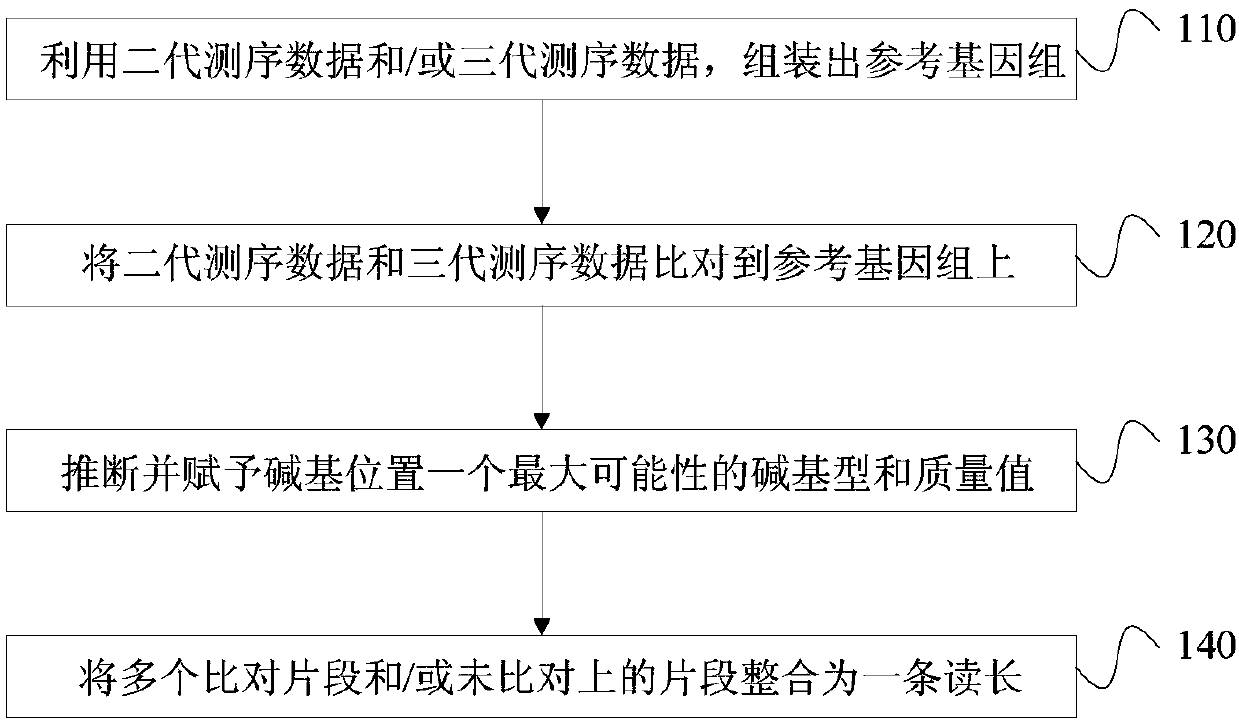
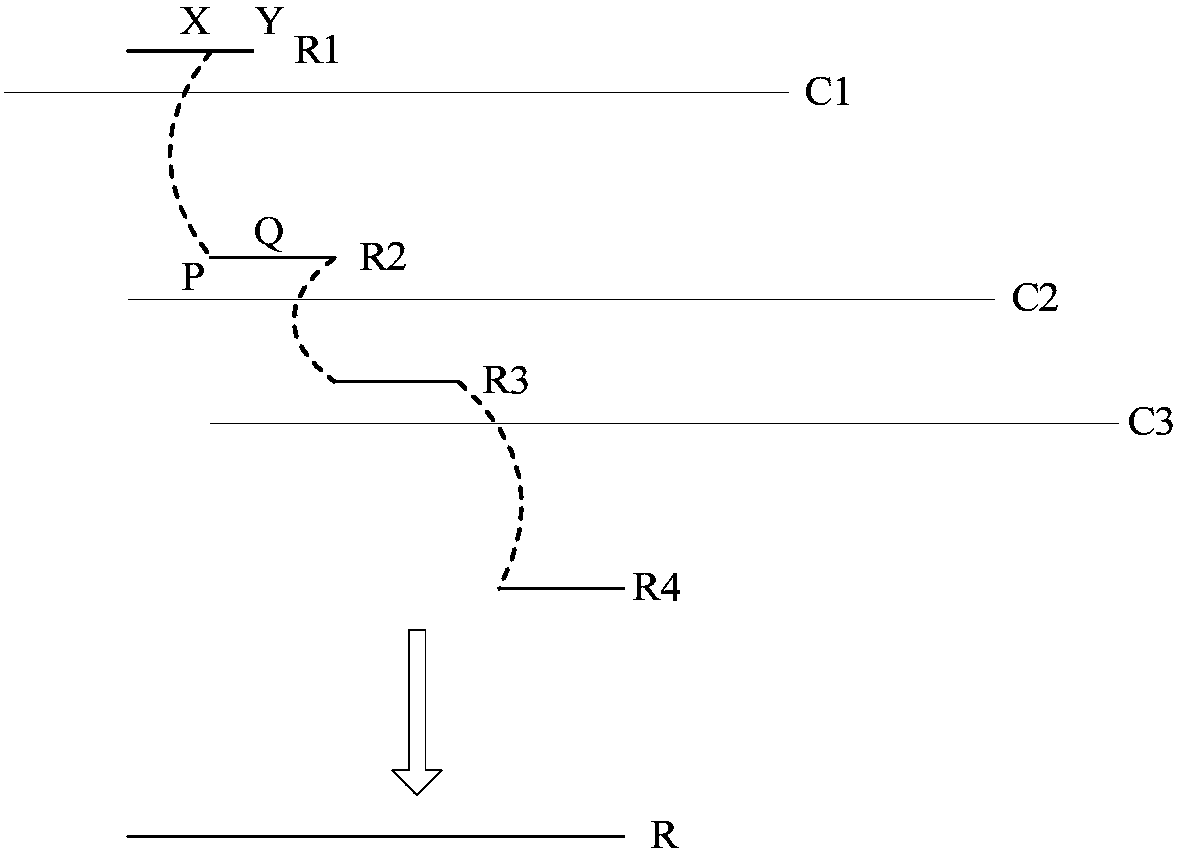
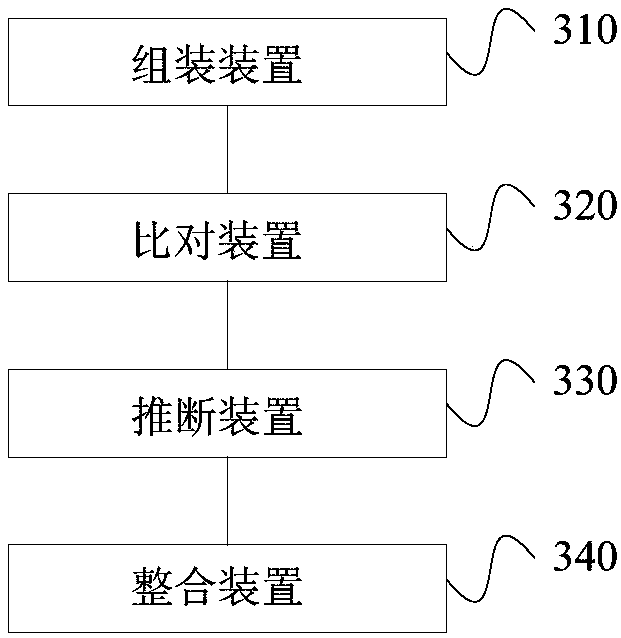
Third-generation sequencing data error correction method, third-generation sequencing data error correction device and computer readable storage medium
A sequencing data and next-generation sequencing technology, applied in the field of bioinformatics, can solve problems such as data loss, inapplicability of low-depth data, and no quality value system
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0082] Example 1: Mixed error correction of human chromosome 22 PacBio sequencing data
[0083] The original data in this example are 68×Hiseq PE250 sequencing data and 90×Pacbio sequencing data. In this embodiment, we extracted 15×Pacbio sequencing data as example data to test the performance of the present invention under low-depth data.
[0084] 1. Using 68×Hiseq PE250 sequencing data and 15×Pacbio sequencing data, use DBG2OLC software for mixed assembly to obtain a preliminary reference genome.
[0085] 2. The statistical results of the data show that the N50 of the assembled contig (contig) is 289kb, and the self-alignment shows that there are no long-segment high-repetition sequences, and the assembly results are suitable for use as a reference sequence for alignment without any processing.
[0086] 3. Use the bwa software to compare the Hiseq sequencing data and the Pacbio sequencing data to the reference sequence.
[0087] 4. Use the maximum a posteriori model to inf...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com