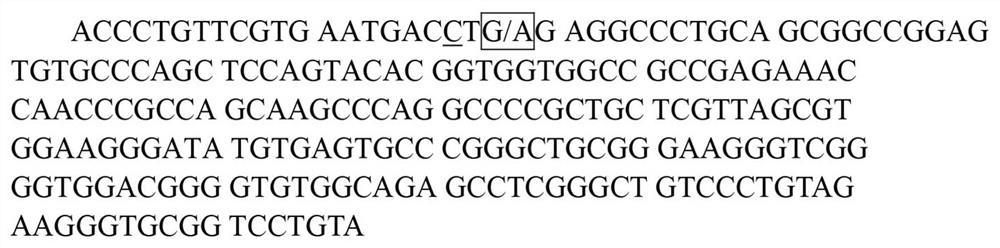Method for detecting single nucleotide polymorphism site of cattle ret gene, detection kit and application thereof
A single nucleotide polymorphism, detection kit technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problem of offspring sickness, inability to screen for HSCR pathogenic mutation sites, household heads or feedlots Losses and other issues, to achieve important effects of economic value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1 Amplification of Cattle RET Gene Sequence and Detection of Polymorphic Sites
[0048] 1.1 Design of PCR primers for cattle RET gene
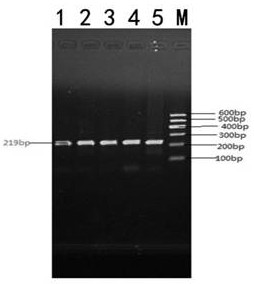
[0049] Taking the bovine RET (AC_000185.1) sequence published by NCBI as a reference, the 219bp gene fragment of the RET gene from 34669bp to 34887bp is as follows figure 1 shown. Use Primer 5.0 to design a PCR primer pair that can amplify the 34690th region of the cattle RET gene. The primer sequences are as follows:
[0050] Upstream primer P3: 5'-TCTCTGGGCTGAGTGGGAAA-3';
[0051] Downstream primer P4: 5'-CAAGGAAGCCCACCAGTCAG-3';
[0052] Using P3 and P4 primers to carry out PCR amplification on the cattle genome, a 486bp gene fragment including the 34690th position of the cattle RET gene (AC_000185.1 sequence) can be amplified. After sequencing and identification of the amplified fragments, after analysis, It was found that there was a SNP polymorphism at position 34690 of the RET gene.
[0053] Since there is no natural...
Embodiment 2
[0112] Example 2 The application of the single nucleotide polymorphism site detection method of yellow cattle RET gene in molecular breeding
[0113] Genotype data: genotypes (AG, GG and AA) recognized by MaeI (XspI).
[0114] Production data: birth weight of Nanyang and Jiaxian Red Cattle, as well as body weight, body height, body oblique length, bust, ischial end width, and daily gain at 6, 12, 18 and 24 months of age.
[0115] Correlation analysis model: First, describe and analyze the data to determine whether there are outliers, and then use the least square method to analyze and correct the data; according to the data characteristics, use SPSS (18.0) software to analyze the effects of production traits among genotypes. A fixed model was used in the analysis of genotype effects:
[0116] Y ijkl =μ+BF i +Month j +G k +e ijkl
[0117] Among them: Y ijkl is the observed value of traits, μ is the overall mean, BF i is the fixed effect of the i-th variety and farm, Mo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com