A dual-mode nucleic acid detection method based on aggregation-induced luminescence/surface plasmon colorimetric analysis
A technology of aggregation-induced luminescence and surface plasmon, which is applied in biochemical equipment and methods, and the determination/inspection of microorganisms. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] (1) H1 and H2 DNA were first dissolved in 1×PBS buffer, and then heated to 95°C, kept at a constant temperature for 5 minutes, and then lowered to room temperature and kept for 1 hour. Then mix H1 and H2 for later use. The sequences of H1 and H2 DNA are:
[0035] DNA-H1: ttaacccacgccgaatcctagactcaaagtagtctaggattcggcgtg (SEQ ID No. 1);
[0036] DNA-H2: agtctaggattcggcgtgggttaacacgccgaatcctagactactttg (SEQ ID No. 2).
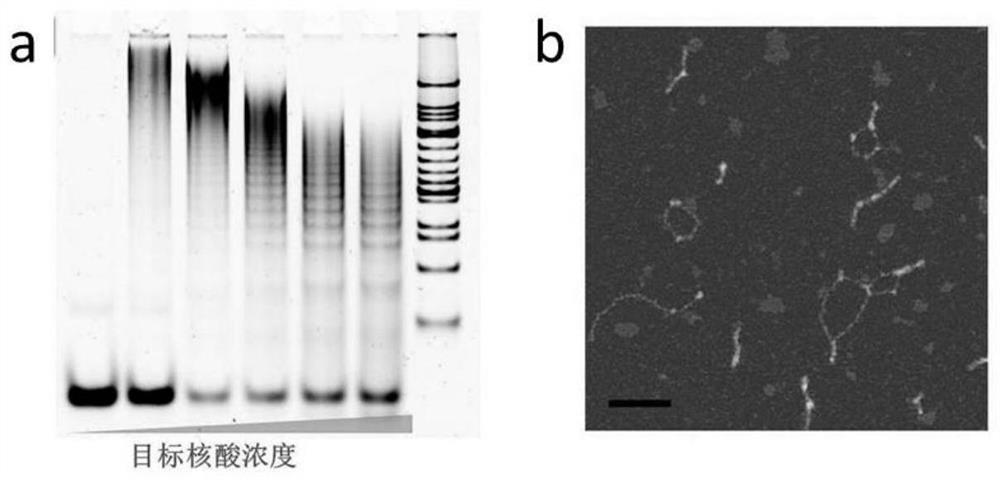
[0037] (2) Different concentrations of target DNA (0.0625 μM, 0.125 μM, 0.25 μM, 0.5 μM and 1 μM) were mixed with H1 and H2, and reacted at room temperature for 2 hours to obtain HCR reaction products. The HCR product was processed by ultrafiltration to reduce background signal. The specific steps are as follows: the HCR product is firstly diluted with 1×PBS with a dilution factor of 4, and then the diluted solution is added to a 100 kDa (purchased from Millipore) ultrafiltration tube. The samples were centrifuged (10000r / min, 2min) and then sonicated f...
Embodiment 2
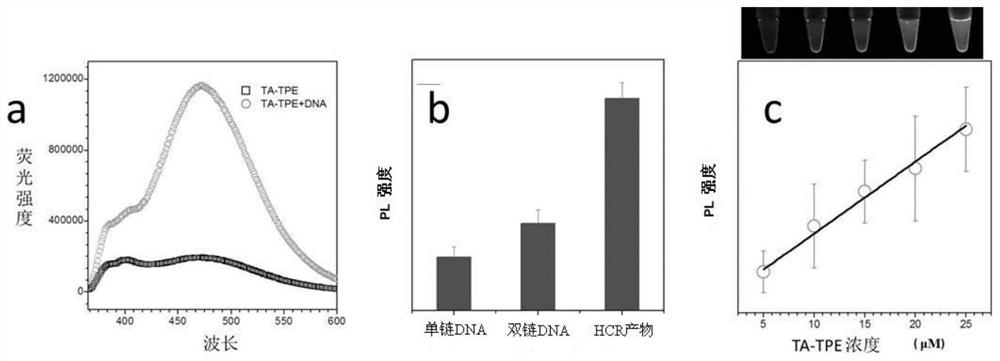
[0053] (1) AuNPs were first incubated overnight with thiol-modified DNA, and the molar concentration ratios of AuNPs to DNA were 1:50 (low), 1:100 (moderate) and 1:300 (high). Then add 10 μL of 1×PBS solution to the solution every half hour for a total of three times. React at room temperature for 7 hours. The samples were then centrifuged and washed three times with deionized water. Finally, the precipitate was resuspended with deionized water to a concentration of 10 nM to obtain a gold nanoparticle solution modified with thiol DNA (the DNA modified on the gold nanoparticle is to improve the stability of the gold nanoparticle, and its sequence can be any sequence).
[0054] (2) Take 3 different molar ratios of sulfhydryl DNA-modified gold nanoparticle solutions, take 100 μL each and add them to a 96-well plate (a total of 6 wells), and add TA-TPE to the final concentrations of 0.5 μmol and 1 μmol respectively , 2μmol, 3μmol, 4μmol, 5μmol. Take pictures to observe the chan...
Embodiment 3
[0057] The detection method for microRNA in this embodiment is similar to the detection method for DNA in Example 1. Different from DNA, the sequences of H1 and H2 used in RNA and detection are as follows:
[0058] RNA sequence: uggagugugacaaugguguuuga (SEQ ID No.10);
[0059] RNA-H1: tggagtgtgacaatggtgtttgaaccattgtcacactccaaattgc (SEQ ID No. 11);
[0060] RNA-H2: tcaaacaccattgtcacactccagcaatttggagtgtgacaatggt (SEQ ID No. 12).
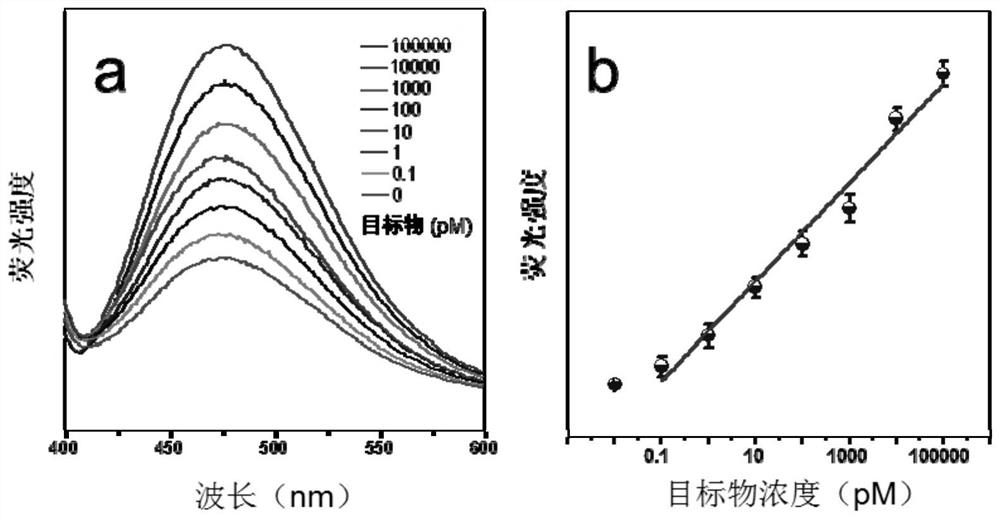
[0061] In this example, the fluorescence spectra of TA-TPE interacted with HCR products of different concentrations of target RNA (target RNA concentrations were 0, 10nM, 20nM, 40nM, 60nM, 80nM, 100nM) were as follows: Figure 8 Shown in a; according to the standard curve of fluorescence intensity and microRNA concentration in a Figure 8 Shown in b. From Figure 8 It can be seen that as the concentration of target RNA increases, the fluorescence intensity also increases, and presents a good linearity.
PUM
| Property | Measurement | Unit |
|---|---|---|
| size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com