Azotobacter chroococcum and identification method and application thereof
A technology of nitrogen-fixing bacteria and strains, which is applied in the field of microorganisms and can solve problems such as long cycles
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] Example 1: Using SINOPEC12 to identify soil samples above oil and gas reservoirs.
[0067] (1) Design specific primers based on the conserved 16S rDNA sequence of Azotobacter terescens SINOPEC12.
[0068] The sequences of the designed specific primers are as follows:
[0069] Upstream primer: 5'-AACTGAGACACGGTCCAGACTCCTACGG-3';
[0070] Downstream primer: 5'-CAACCCTCTGTACCGACCATTGTAGCAC-3';
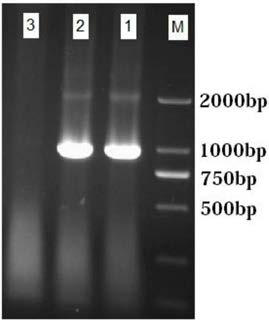
[0071] The size of the target band amplified by the specific primer is about 1000bp.
[0072] (2) Extract the whole genome of microorganisms in the soil.
[0073] Weigh 0.5g of the soil above the oil and gas reservoir and the background soil respectively, add them to the Lysing Matrix E Tube, and use a cell disruptor to break the cells (speed 4.5m / s, 30s, 4 times), and the broken cells according to SPIN Kit for Soil (USA MP Biomedicals biomedical company product) specification sheet carries out follow-up operation, obtains the complete genome of microorganism in the soil samp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com