Method and kit for the generation of DNA libraries for massively parallel sequencing
A parallel sequencing, large-scale technology, used in DNA preparation, recombinant DNA technology, biochemical equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
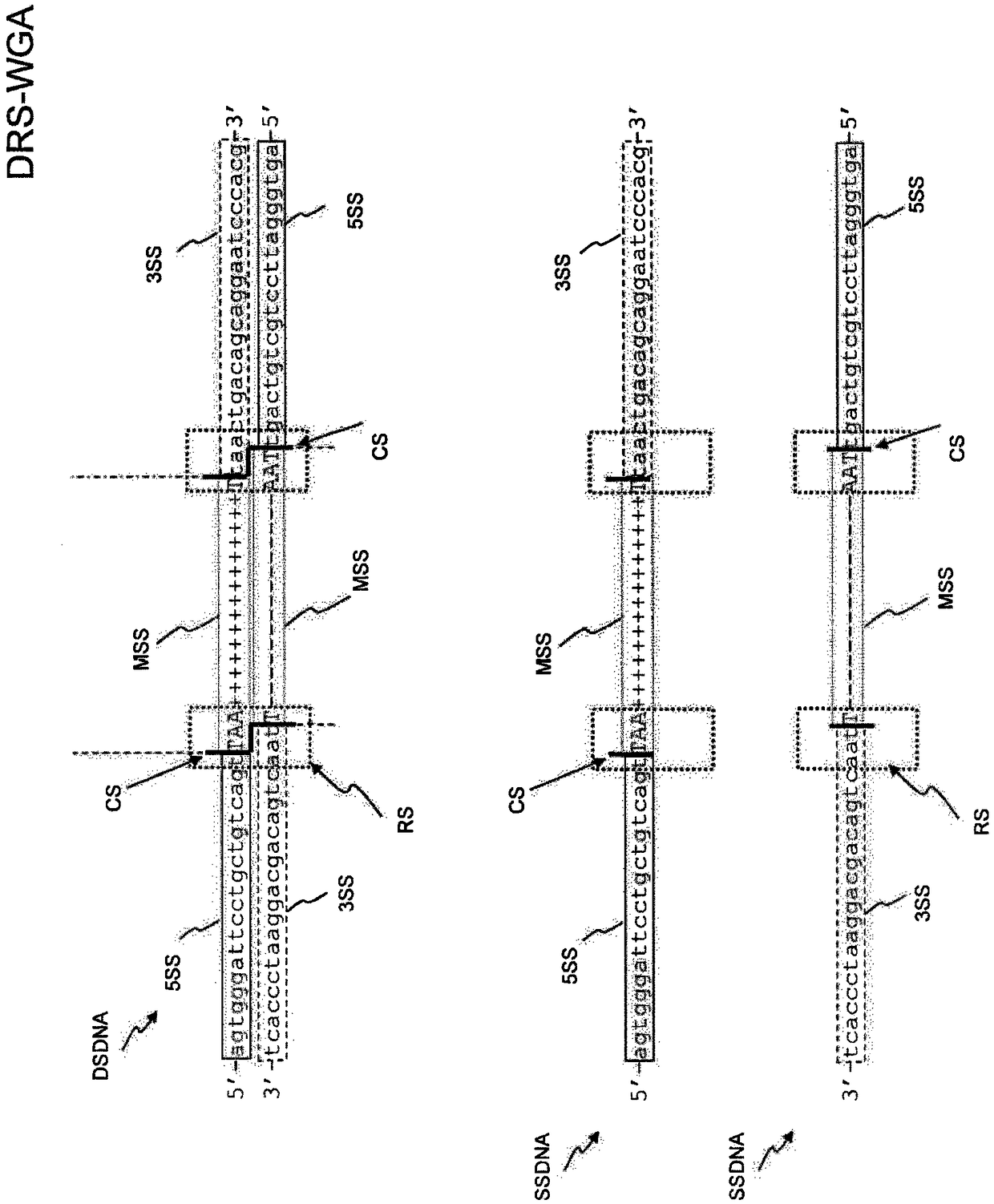
[0153] Scheme of LPWGS on Ion Torrent PGM after DRS-WGA
[0154] 1) Definitive restriction site whole genome amplification (DRS-WGA)
[0155] According to the manufacturer's instructions, use Ampli1 TM The WGA kit (Silicon Biosystems) amplifies single-cell DNA.
[0156] Ampli1 TM The WGA kit is designed to provide whole genome amplification from DNA obtained from a single cell. After cell lysis, the DNA is digested with a restriction enzyme, preferably MseI, and universal adapter sequences are ligated to the DNA fragments. Amplification is mediated by a single specific PCR primer for all generated fragments ranging in length from 200-1,000 bp distributed throughout the genome.
[0157] 2) Re-amplification of WGA products
[0158] 5 μL of WGA-amplified DNA was diluted by adding 5 μL of nuclease-free water and purified using the Agencourt AMPureXP system (Beckman Coulter) to remove unbound oligonucleotides and excess nucleotides, salts and enzymes.
[0159] Bead-based DNA...
Embodiment 2
[0200] Prototype for LPWGS on Ion Torrent Proton after DRS-WGA
[0201] 1. Definitive restriction site whole genome amplification (DRS-WGA)
[0202] Ampli1 was used according to the manufacturer's instructions as detailed in the previous example TM Single-cell DNA was amplified with the WGA kit (Menarini Silicon Biosystems).
[0203] 2. Double-Stranded DNA Synthesis
[0204] According to the manufacturing scheme, using Ampli1 TM The ReAmp / ds kit converts 5 μL of WGA-amplified DNA into double-stranded DNA (dsDNA). This process ensures the conversion of single-stranded DNA (ssDNA) molecules into dsDNA molecules.
[0205] 3. Purification of dsDNA products
[0206] 44 μL of nuclease-free water was added to dilute 6 μL of dsDNA synthesis product and purified by Agencourt AMPure XP beads (Beckman Coulter) to remove unbound oligonucleotides and excess nucleotides, salts and enzymes. Bead-based DNA purification was performed according to the following protocol: 75 μL (ratio: 1...
Embodiment 3
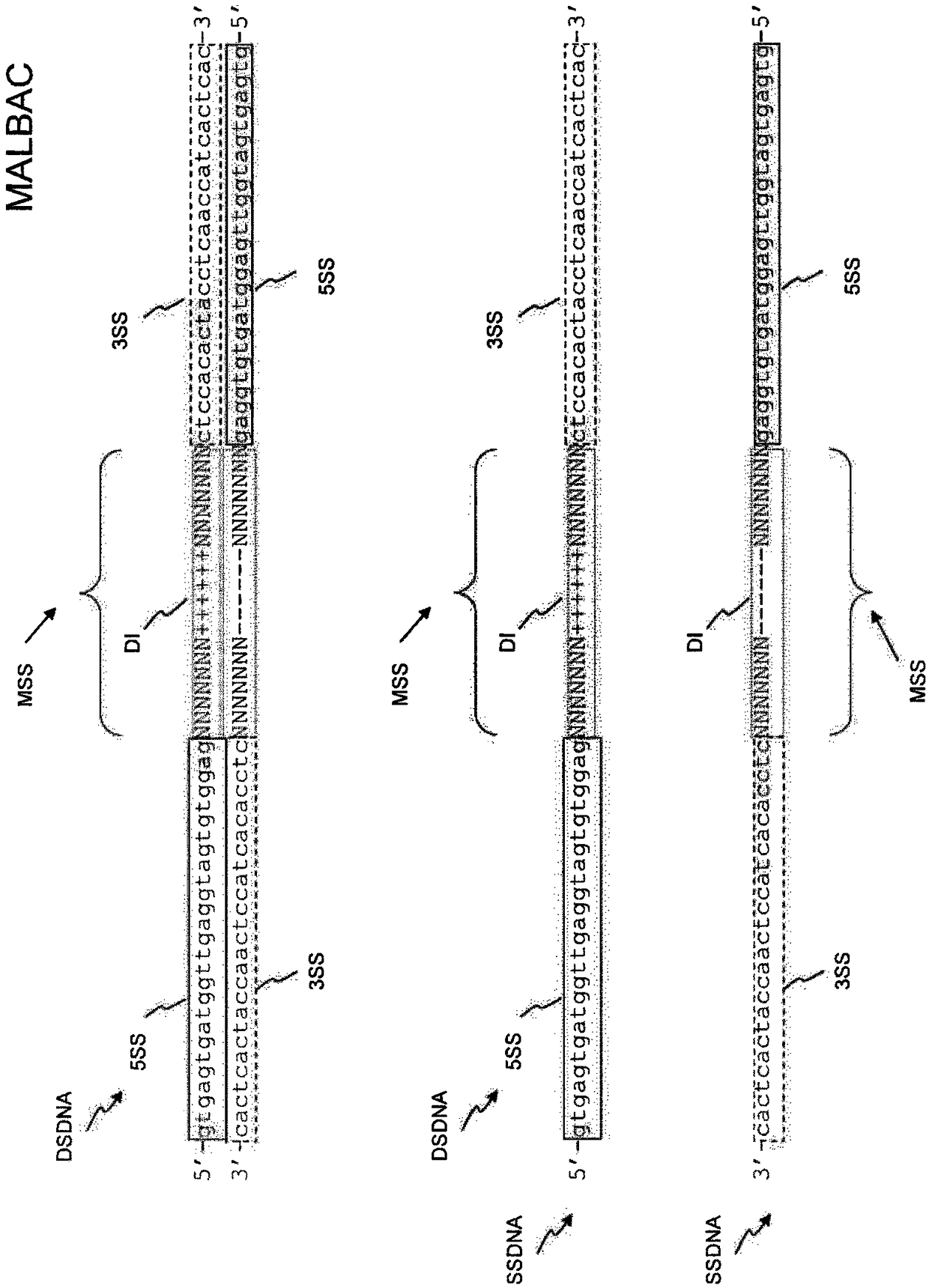
[0227] Protocol for Low-Pass Whole-Genome Sequencing on the Illumina MiSeq
[0228] plan 1
[0229] Definitive restriction site whole genome amplification (DRS-WGA):
[0230] According to the manufacturer's instructions, use Ampli1 TM The WGA kit (Silicon Biosystems) amplifies single-cell DNA. 5 μL of WGA-amplified DNA was diluted by adding 5 μL of nuclease-free water and purified using the Agencourt AMPure XP system (scale 1.8×). DNA was eluted in 12.5 μL and quantified by the dsDNA HS assay on a Qubit 2.0 fluorometer.
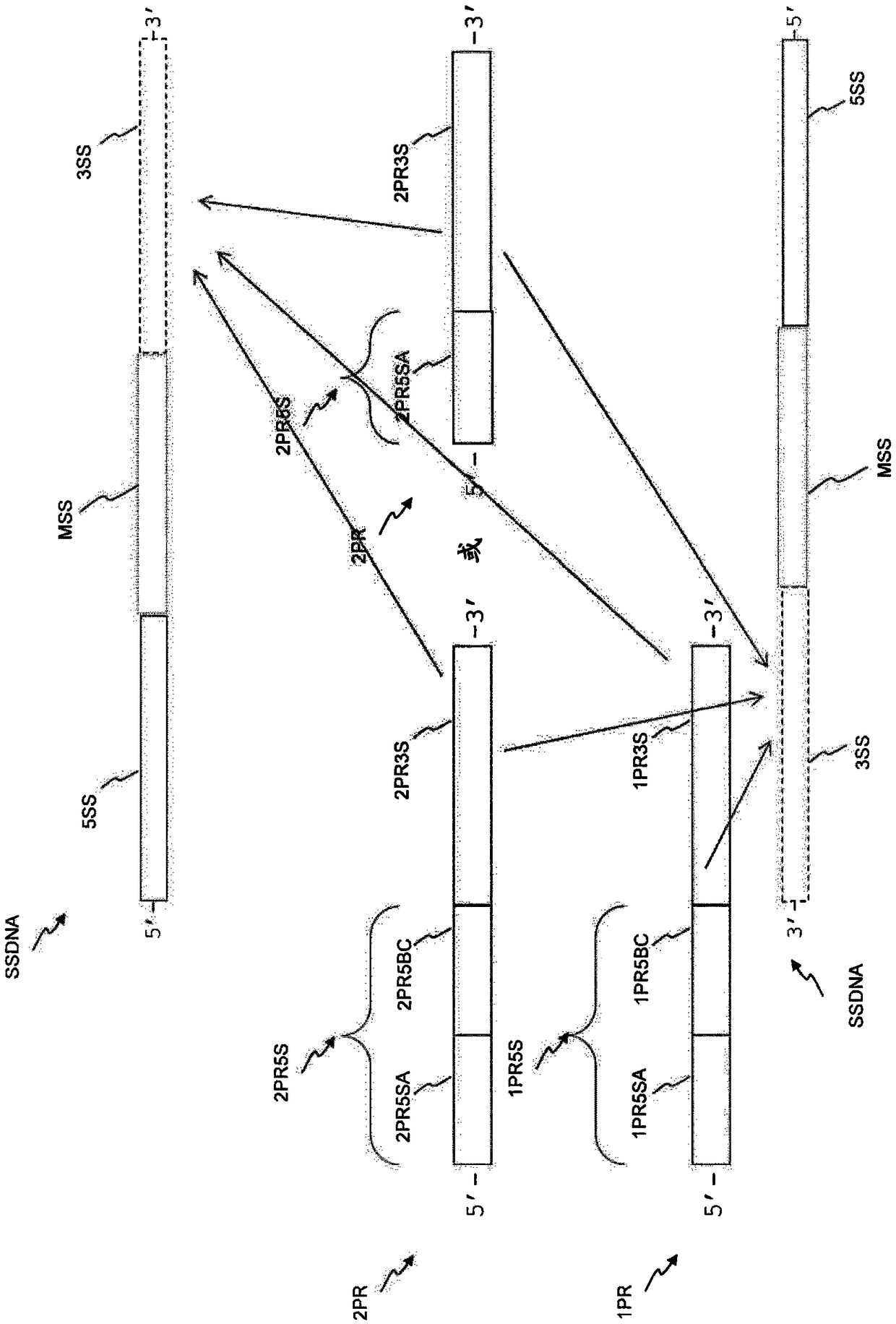
[0231] barcoded reamplification
[0232] Such as Figure 4 is schematically shown in, using Ampli1 TM Barcoding reamplification was performed with a PCR kit (Menarini Silicon Biosystems) in a volume of 50 μl. Each PCR reaction was composed as follows: 5 μl Ampli1 TM PCR reaction buffer (10×), 1 μL of one primer (25 μM) in SEQ ID NO: 195 to SEQ ID NO: 202, 1 μl of one primer (25 μM) in SEQ ID NO: 203 to SEQ ID NO: 214 primers, 1.75 μl Ampli1 TM ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com