Detection method and system for colorectal cancer
A colorectal cancer, target nucleic acid technology, applied in biochemical equipment and methods, microbial determination/examination, DNA/RNA fragments, etc., can solve the problems of low diagnostic value and large trauma of carcinoembryonic antigen
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1 Screening and verification of biomarkers
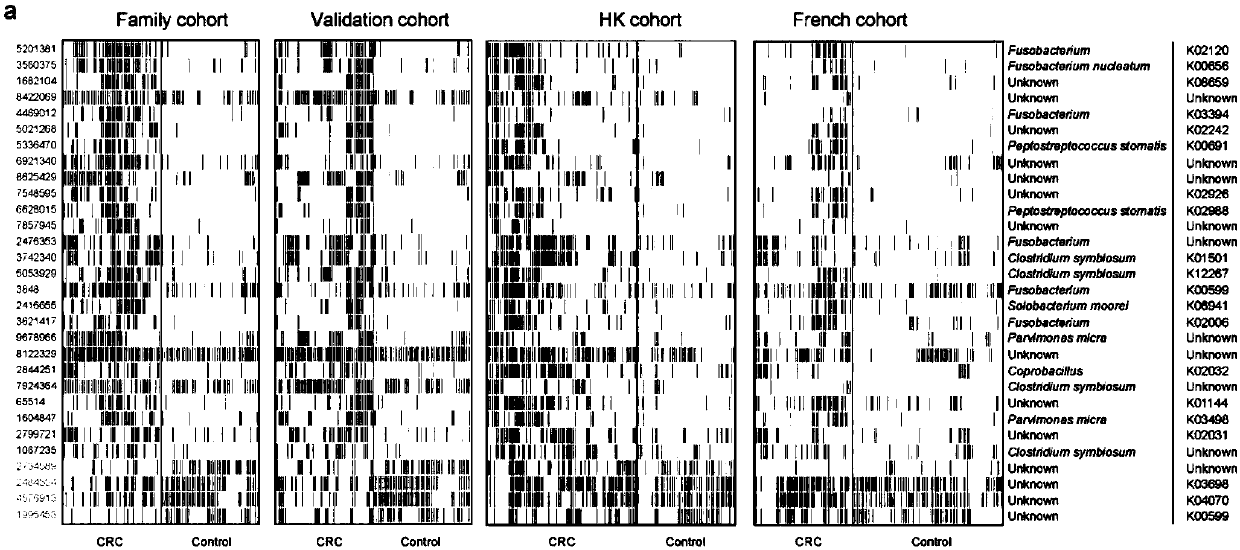
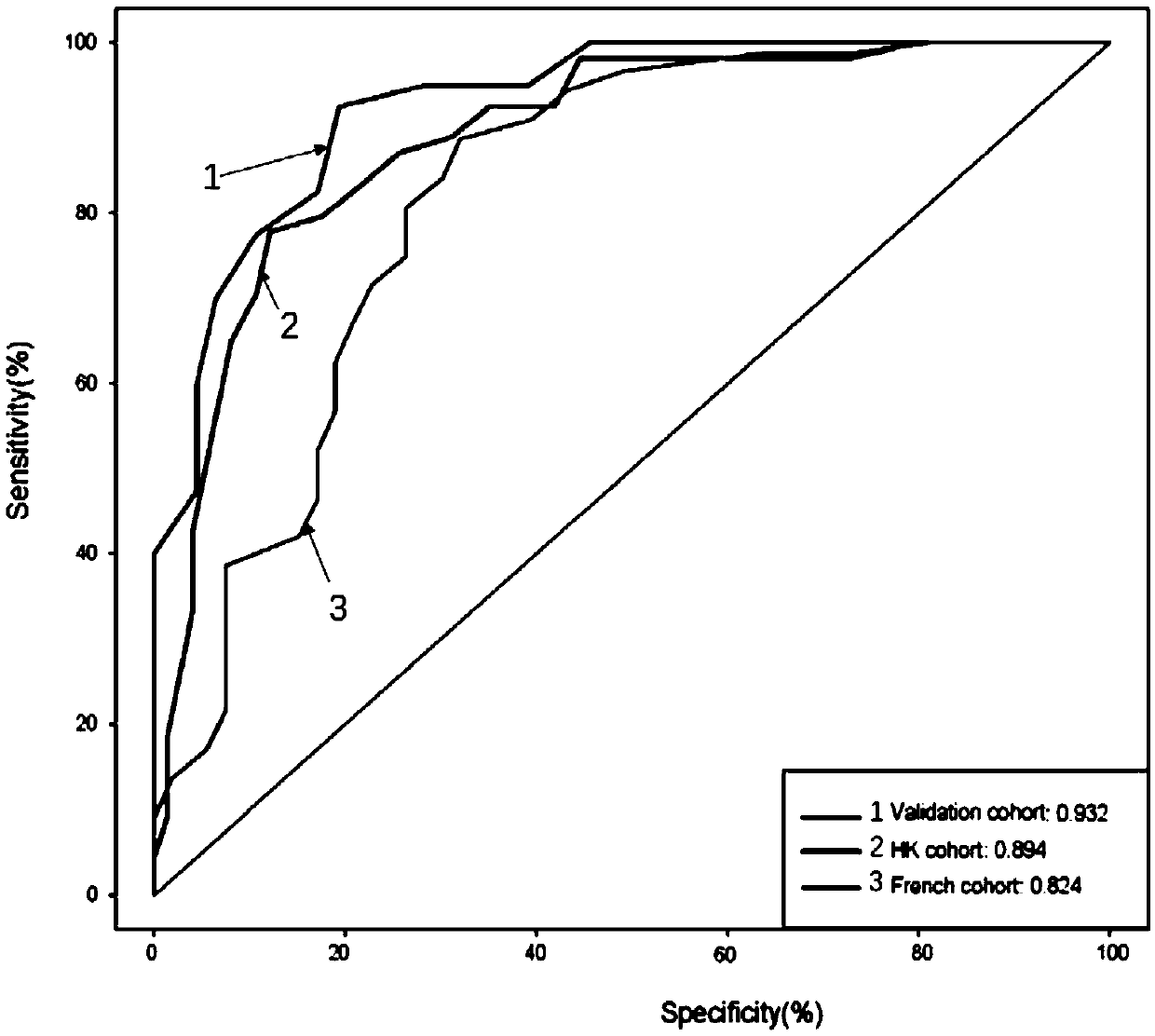
[0050] Collected 193 stool samples from Southwest Hospital in Chongqing, China, including 98 colorectal cancer patient samples (68 male samples, 30 female samples) and 95 healthy control samples (49 male samples, 46 female samples) . We performed metagenomic detection on 52 colon cancer patient samples and 55 healthy control samples (from healthy family members with similar diet structure, living habits, and living environment) to detect colorectal cancer Differences in gut microbiota between patients and healthy people. In the process of detecting the above samples, we identified 30 microbial gene fragments related to colorectal cancer, of which 26 gene fragments were increased in colorectal cancer patient samples, and 4 gene fragments were increased in healthy control samples increased in (see figure 1 ). figure 2 The receiver operating characteristic (ROC) curve showed that the 30 candidate gene fragments show...
Embodiment 2
[0067] Example 2 Primer Screening
[0068] Specific primers were designed for the biomarker-specific sequences obtained through the screening in Example 1, and primer tests were performed on the specific primers designed for each specific sequence: that is, by ordinary PCR amplification, and the strips were detected by agarose gel electrophoresis. If the size of the target band is the same as expected and the band is single, it indicates that the specificity of the primer is good. After detection, it was found that the primers shown in Table 2 can be used to amplify the specific sequence screened in Example 1, the target band size is consistent with the expectation and the band is single, and the specificity of amplification is good.
[0069] Table 2 Specific primer sequences
[0070]
Embodiment 3
[0071] Embodiment 3 colorectal cancer detection application
[0072]This example is mainly based on the real-time fluorescent quantitative PCR technology to test the DNA of the fecal microbial flora of colorectal cancer patients and normal people, and the results shown are the results provided by the best primers provided in Example 2.
[0073] 1. Sample collection
[0074] Sampling swabs were used to collect the central parts of feces from 30 colorectal cancer patients and 30 normal people, and stored in the feces collection tube containing buffer solution. After tightening the tube cap, turn it upside down 5 or 6 times, and mix the preservation solution and The feces are well mixed.
[0075] Specimen storage and transportation: store at room temperature or refrigerated at 4°C. Specimens can be transported at room temperature for long-distance transport before refrigeration, and dry ice is required for long-distance transport of specimens after refrigeration.
[0076] 2. N...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com