Colorectal cancer marker and application thereof
A technology of colorectal cancer and markers, which is applied in the fields of colorectal cancer markers, kits, colorectal cancer detection, primer pairs, and nucleic acid probes. And other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] The MGWAS method was used to search for markers of colorectal cancer intestinal flora (this method is well known in the art and described in detail in the literature), and the specific sequence of Fusobacterium nucleatum was found, as shown in Table 1 below. Among them, the MGWAS (genome-wide association analysis) method uses natural populations and artificial populations as research materials, and conducts genome-wide association analysis of metabolomes based on resequencing information and combined with metabolome results. References for the experimental process: Jun Yu, QiangFeng eal.,. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 2015; 0:1–9. doi:10.1136 / gutjnl-2015-309800, according to It needs to be incorporated in whole or in part into this application.
[0043] Table 1 Specific gene sequence
[0044]
[0045]
[0046]
[0047]
[0048] Wherein, the base N in the sequence SEQ ...
Embodiment 2
[0050] The oligo7 software was used to design primers for the specific sequences of the specific bacterial groups screened. On the basis of meeting the important principles as much as possible, the amplification products of the upper and lower primers were required to be 70bp-200bp, and it was estimated that 3-5 sets of primers were designed for each gene.
[0051] For the specific sequences obtained by screening in Example 1, the primer sequences shown below were designed for the specific amplification of each specific sequence, and the designed primer sequences are shown in the following table:
[0052] Table 2 Primer Sequence
[0053]
[0054]
[0055] For the primers that are obtained by design, carry out primer test: promptly utilize common PCR to amplify, agarose gel electrophoresis detects the amplified band, finds through testing: the purpose band size is consistent with expectation and band is single, illustrates and utilizes the present invention to implement T...
Embodiment 3
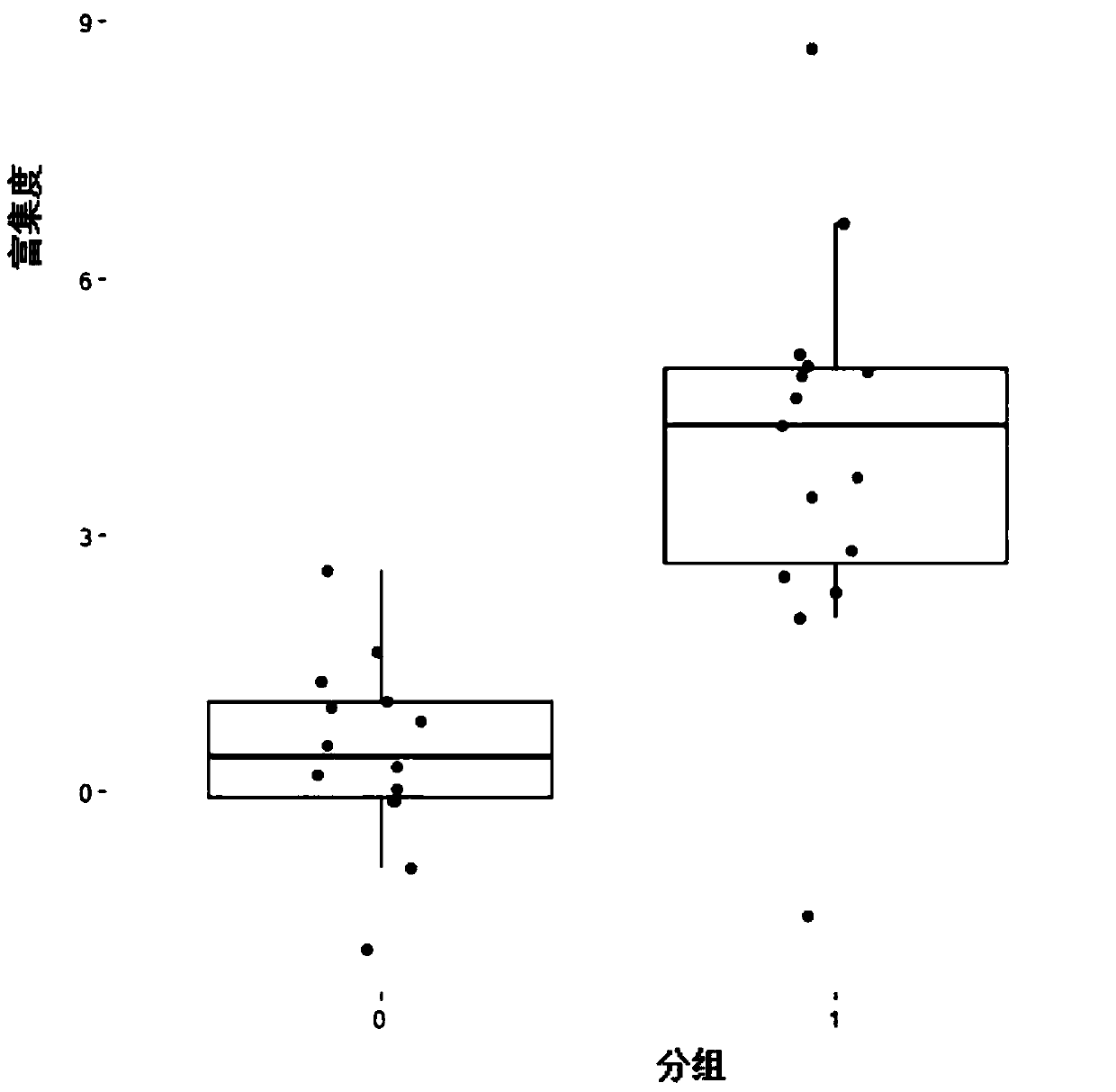
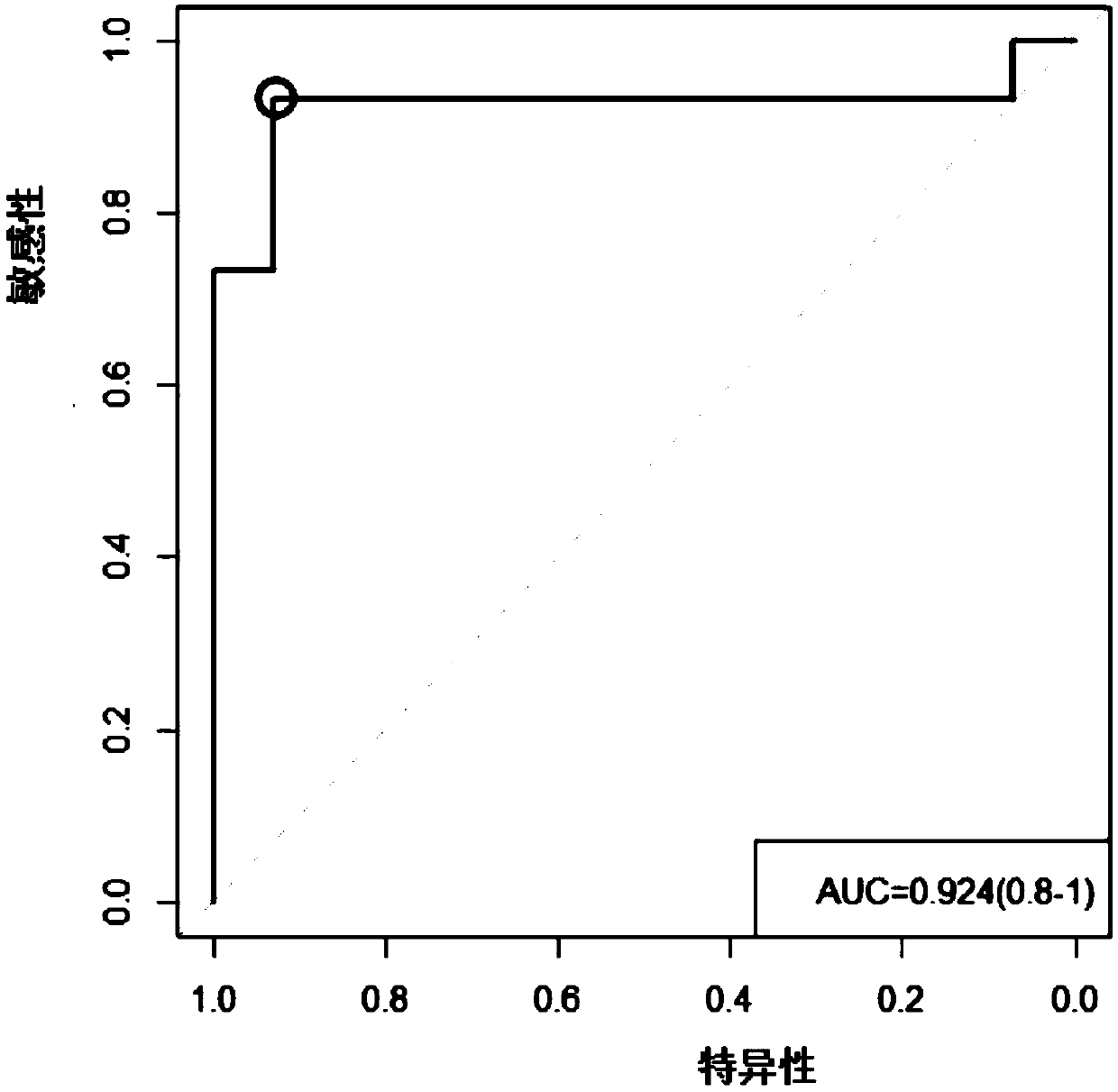
[0057] The embodiment of the present invention is mainly based on the QPCR technology to test the fecal DNA of colorectal cancer patients and normal people, and the results shown are only the results provided by the best primers provided in Example 2. The stool DNA of 16 colorectal cancer patients and 16 normal people were used for QPCR detection (the reagent used was: SYBR Premix Ex Taq(TM) II (Perfect Real Time), product number: DRR081A). Among them, SYBR Premix Ex Taq(TM) II is a special reagent for Real Time PCR (real-time fluorescent quantitative PCR) using the SYBRGreen I mosaic method. Double-stranded DNA is generated by PCR reaction, and SYBR Green I can combine with double-stranded DNA to emit fluorescence. By detecting the intensity of the fluorescent signal in the PCR reaction solution, the target gene can be accurately quantified. Specifically, in the PCR reaction system, an excess of SYBR fluorescent dye is added, and after the SYBR fluorescent dye is specifically...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com