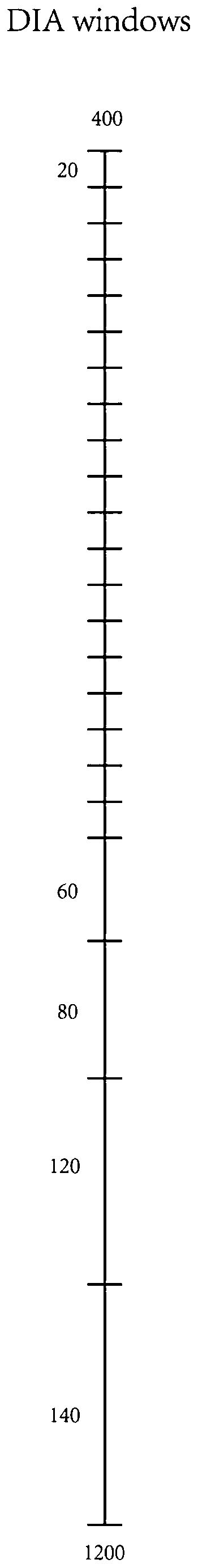A Method for Proteome Detection by Pulsed Data-Independent Acquisition Mass Spectrometry
A mass spectrometry detection and proteome technology, which is applied in the field of pulsed data independent acquisition mass spectrometry detection proteome, can solve the problems of few protein species, low peptide resolution, insufficient resolution, etc., and achieves increased protein quantity, high quality, Reduce the effect of mutual influence
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] The specific technical scheme of the pulse-type data-independent acquisition mass spectrometry detection proteome method provided in this embodiment is as follows:
[0021] A method for detecting proteomes by pulsed data-independent acquisition mass spectrometry, comprising the following steps:
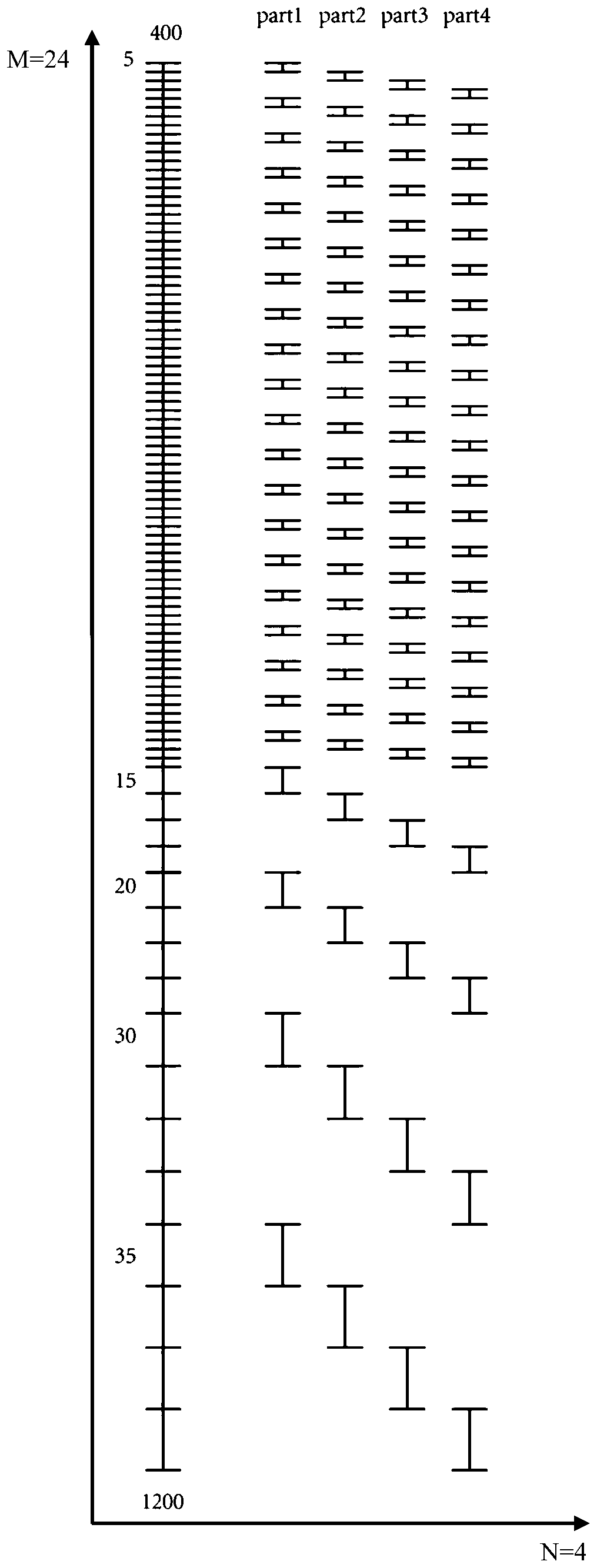
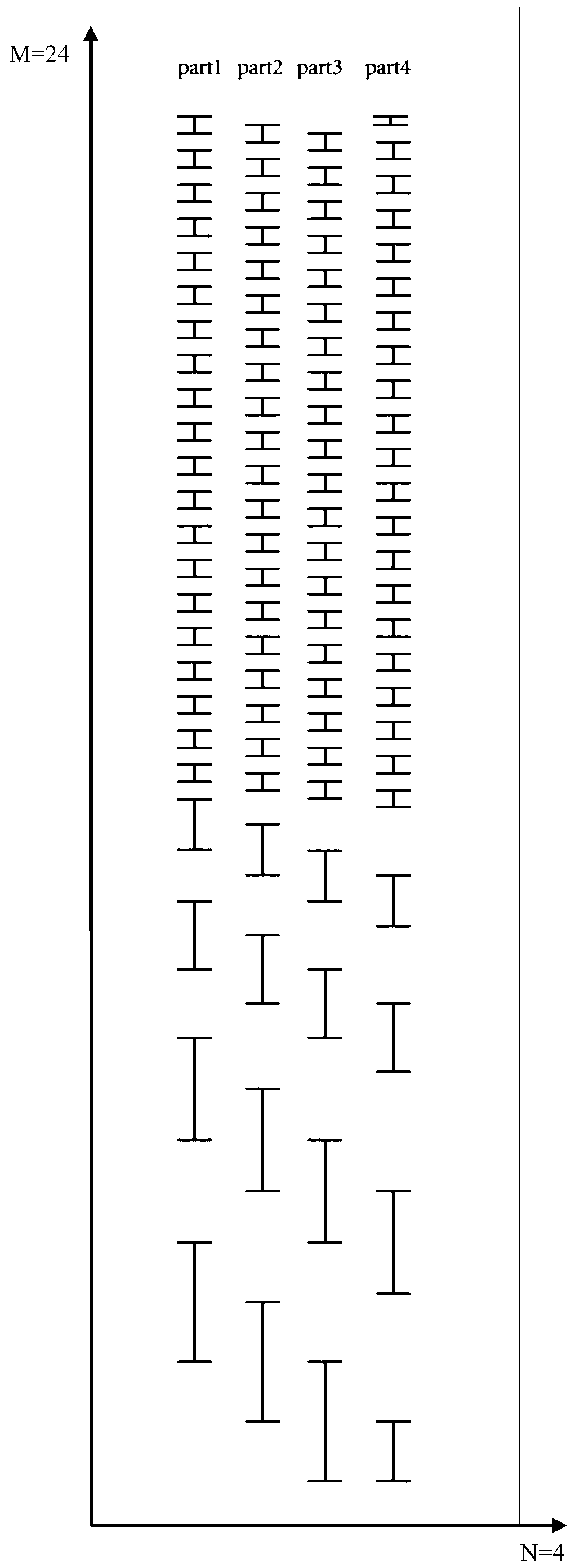
[0022] (1) Set 24 large windows, the interval width of the mass-to-charge ratio of the first 20 large windows is 20m / z, and the interval width of the mass-to-charge ratio of the last 4 large windows is 60m / z, 80m / z, 120m / z z and 140m / z;
[0023] (2) Divide the large window in step (1) evenly into 4 small windows to form 96 small windows, among which, the mass-to-charge ratio interval width of the first 80 small windows is 5m / z, and the last 16 small windows The interval width of the mass-to-charge ratio is 4 15m / z, 4 20m / z, 4 30m / z and 4 35m / z;
[0024] (3) Evenly distribute the 96 small windows in step (2) to 4 mass spectrometry acquisition methods for scanning; for each met...
Embodiment 2
[0028] The specific technical scheme of the pulse-type data-independent acquisition mass spectrometry detection proteome method provided in this embodiment is as follows:
[0029] A method for detecting proteomes by pulsed data-independent acquisition mass spectrometry, comprising the following steps:
[0030] (1) Set 24 large windows, the interval width of the mass-to-charge ratio of the first 20 large windows is 20m / z, and the interval width of the mass-to-charge ratio of the last 4 large windows is 60m / z, 80m / z, 120m / z z and 140m / z;
[0031] (2) Divide the large window in step (1) evenly into 4 small windows to form 96 small windows, of which the interval width of the mass-to-charge ratio of the first 80 small windows is 10 m / z, and the mass-to-charge ratio of the last 16 small windows is The interval width of charge ratio is 4 30m / z, 4 40m / z, 4 60m / z and 4 70m / z;
[0032] (3) Evenly distribute the 96 small windows in step (2) to 4 mass spectrometry acquisition methods fo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com