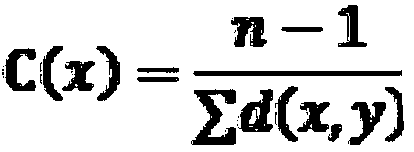Predicting method of nucleic acid binding site bound with protein or micromolecule
A prediction method and binding site technology, applied in proteomics, instrumentation, genomics, etc., can solve the problems of low prediction success rate and multiple false positive results, and achieve the goal of simple method, convenient use, and improved prediction success rate. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1 nucleic acid-small molecule binding site test set
[0050] There are 22 nucleic acid-small molecule complex structures with a length of 20-94 nucleotides in the test set. The information is shown in Table 1.
[0051] Table 1. Test set information of nucleic acid-small molecule complex experimental structure and the positive predictive value of the method of the present invention
[0052]
[0053]
[0054]
[0055] (1) Nucleic acid structure selection
[0056] Download the experimental tertiary structure of nucleic acid-small molecule complexes in the RCSB PDB database as the initial structure information. Take 1EHT as an example, the nucleic acid is the A chain in the 1EHT.pdb file, with 33 bases, the small molecule is TEP, and its molecular formula is C 7 H 8 N 4 O 2 . Then use RNAComposer structure prediction method to predict the model structure of this nucleic acid. In the same way, the experimental tertiary structure and model structure of the remaining 21 nucl...
Embodiment 2
[0075] Example 2 nucleic acid-protein binding site test set
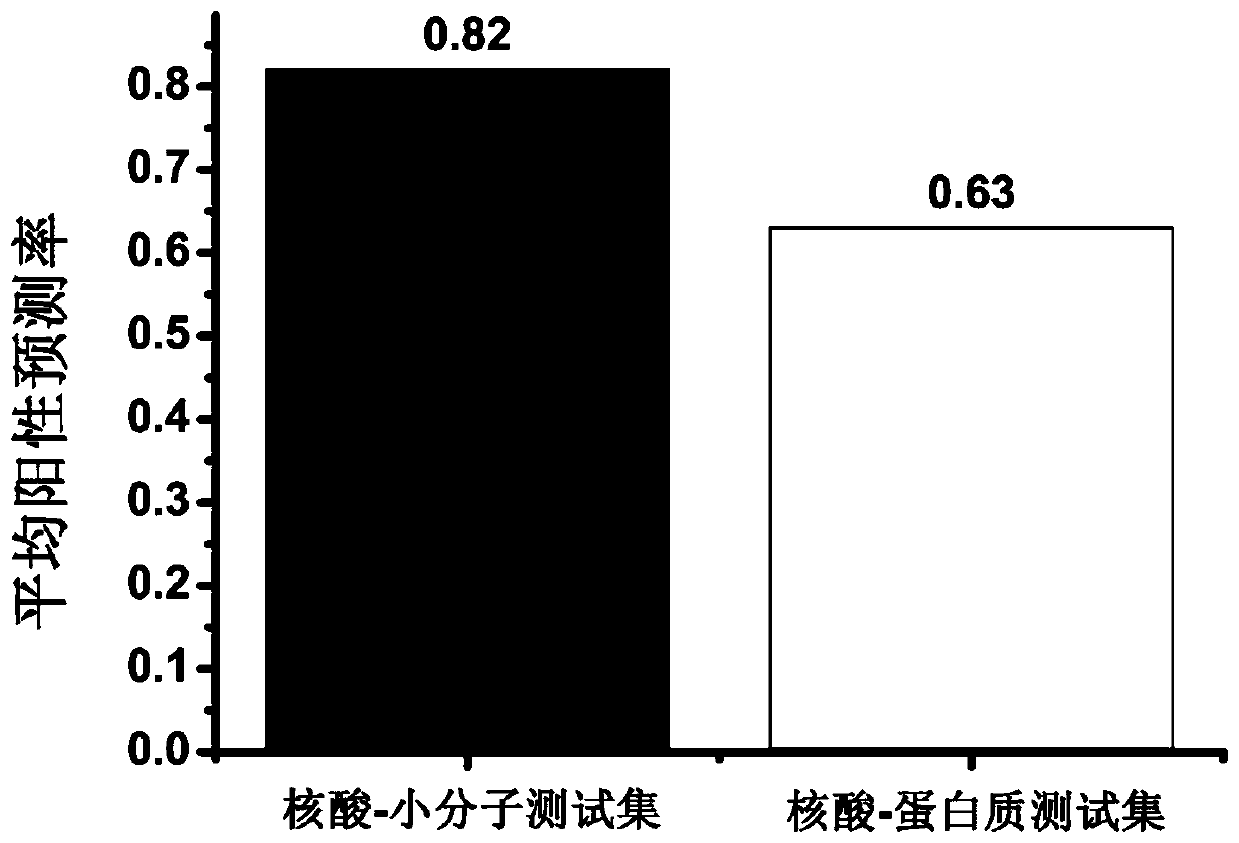
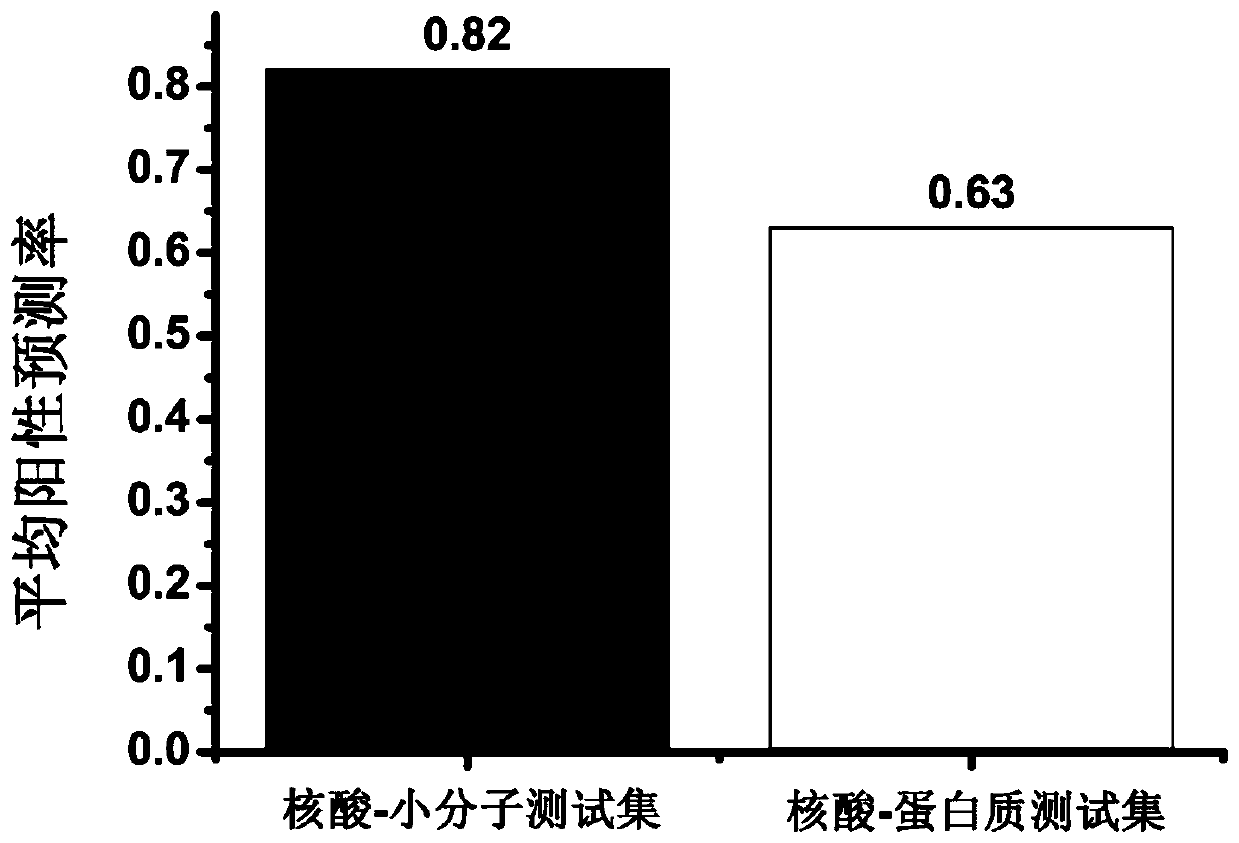
[0076] The test set consists of 72 nucleic acid-protein complex structures, and the length of the nucleic acid ranges from 20 to 157 nucleotides. The information is shown in Table 4. Replace 22 nucleic acid-small molecule complex structures with 72 nucleic acid-protein complex structures, and repeat all the steps in Example 1. The results show that when using the experimental three-level structure, the prediction success rate of the method of the present invention is 63% (Table 4, attached) figure 1 ). When using the model structure (with 25 nucleic acid-protein complex structures removed, and 47 nucleic acid-protein complex structures remaining), the results show that the average positive predictive value of the method of the present invention, Rsite and Rsite2 methods are 66%, 58% and 56% (Table 5), indicating that the nucleic acid binding site prediction accuracy of the present invention is higher than that of mai...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com