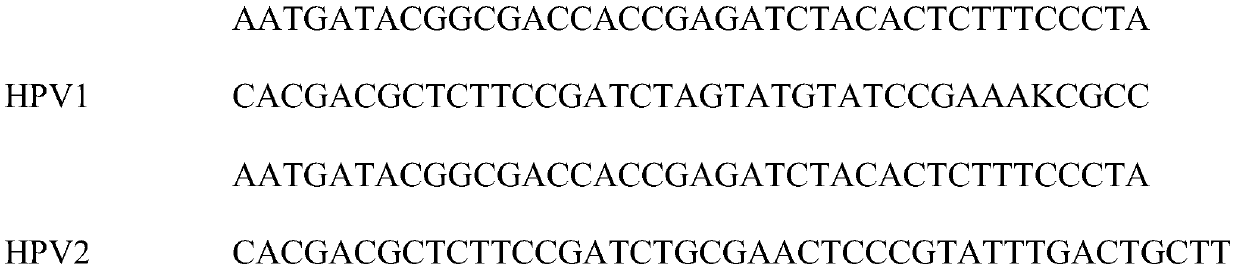Amplification and sequencing method for identification of various HPV types and genomic integration analysis
A technology of integrated analysis and typing identification, applied in the field of genetic diagnosis, can solve the problems of cell canceration, random location, and inability to obtain typing results at the same time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0055] In order to further understand the content of the present invention, the present invention will be described in detail below in conjunction with specific examples.
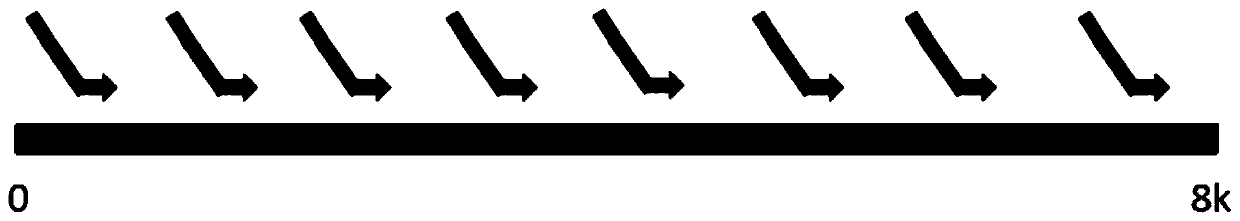
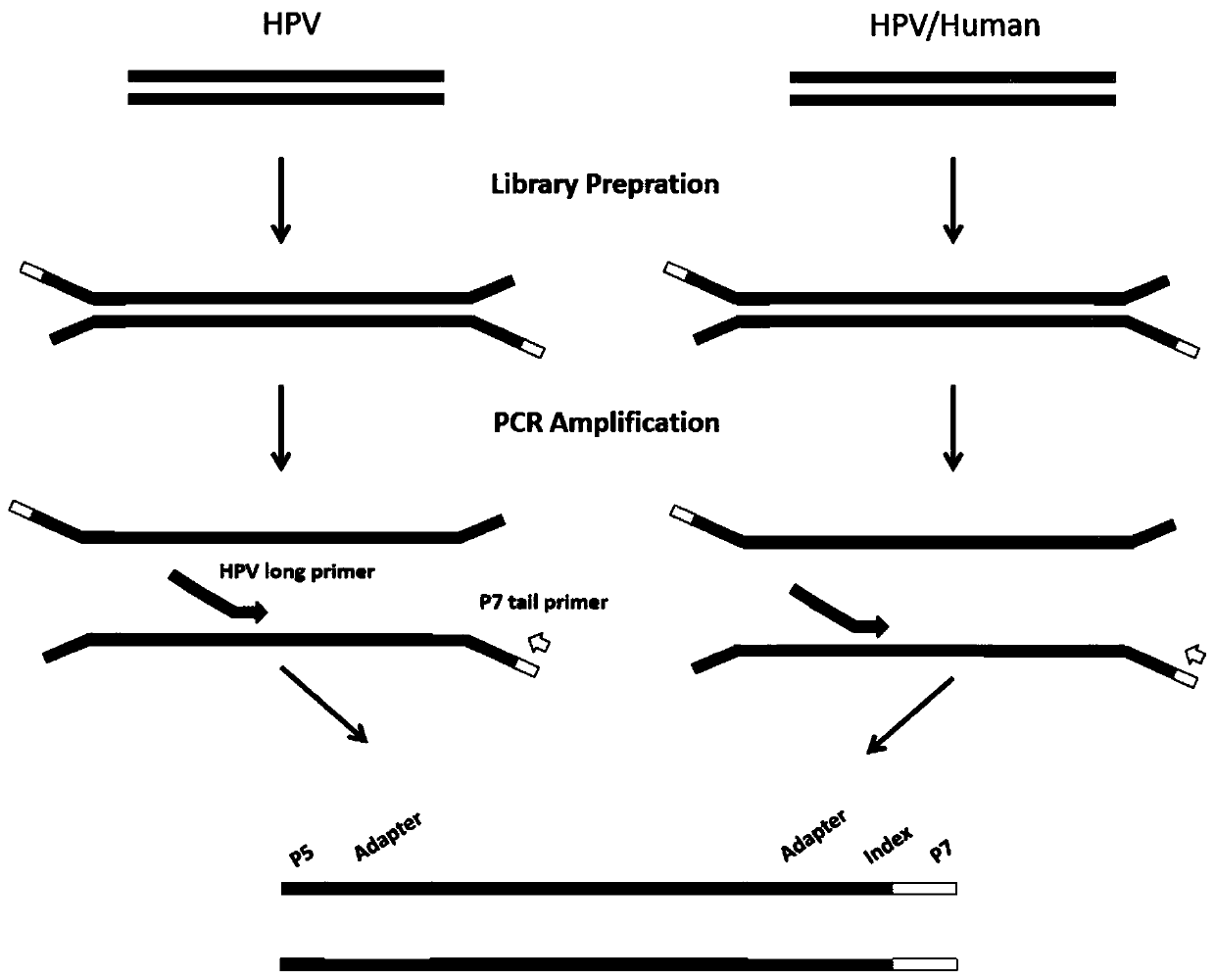
[0056] like figure 1 and figure 2 As shown, there are 29 long primers designed by the present invention, and the long primers are divided into three parts: P5, linker sequence and HPV primers, all of which are forward primers, designed according to the conserved sequences of various HPV types are distributed on the entire 8k HPV genome, and the longest distance between the two primers does not exceed 1k. The first step of the present invention is to interrupt the DNA. The DNA after interruption is screened and purified by magnetic beads, and the main peak is kept at about 350bp fragments, then normal end repair and ligation plus adapters, magnetic bead purification once, and finally PCR amplification enrichment, using the library construction product in the previous step as a template, 29 primer mixtures ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com