High-throughput absolute quantification method for three-domain microorganisms in soil
An absolute quantitative and microbiological technology, applied in the field of microbiology, can solve problems such as wrong conclusions and difficult realization of domain collapse, and achieve the effect of simple experimental operation and favorable follow-up analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
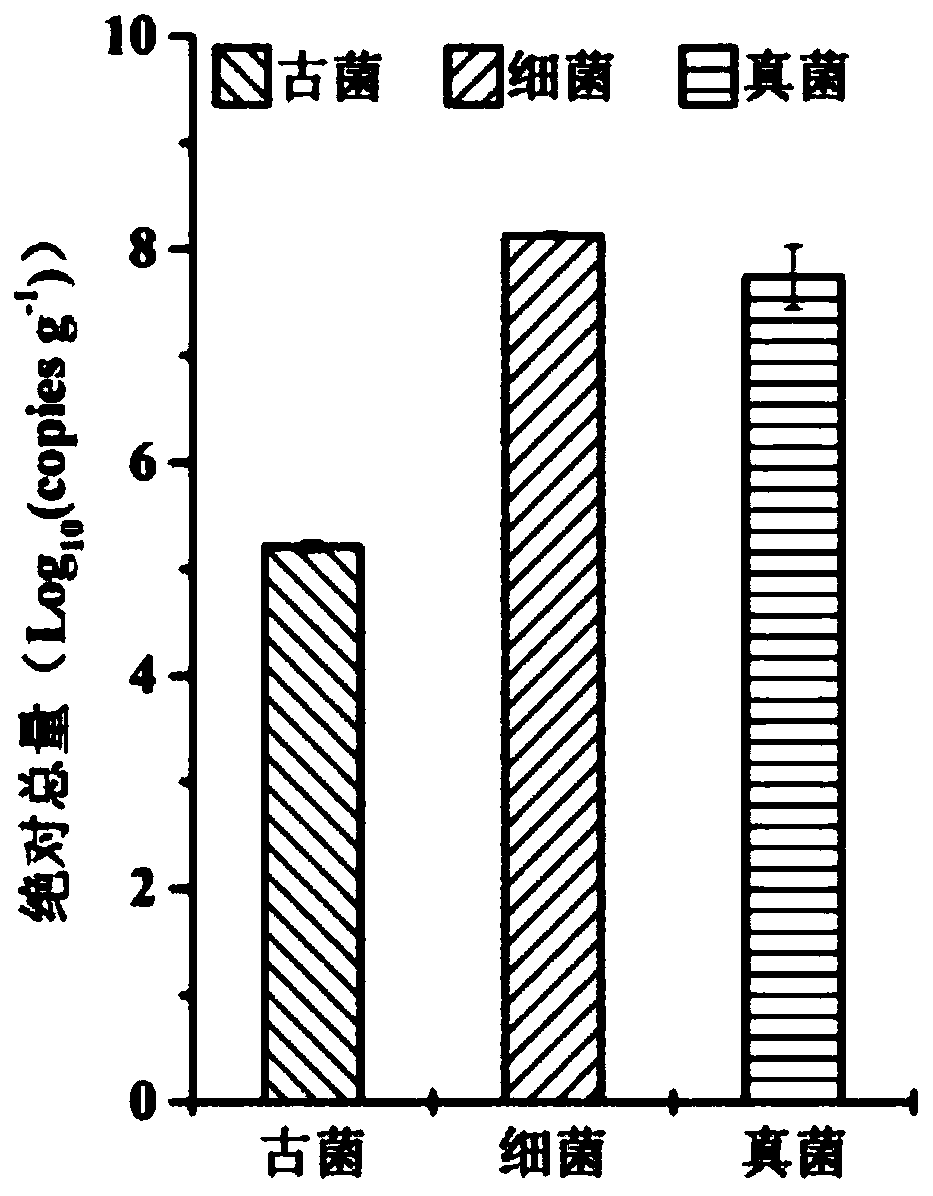
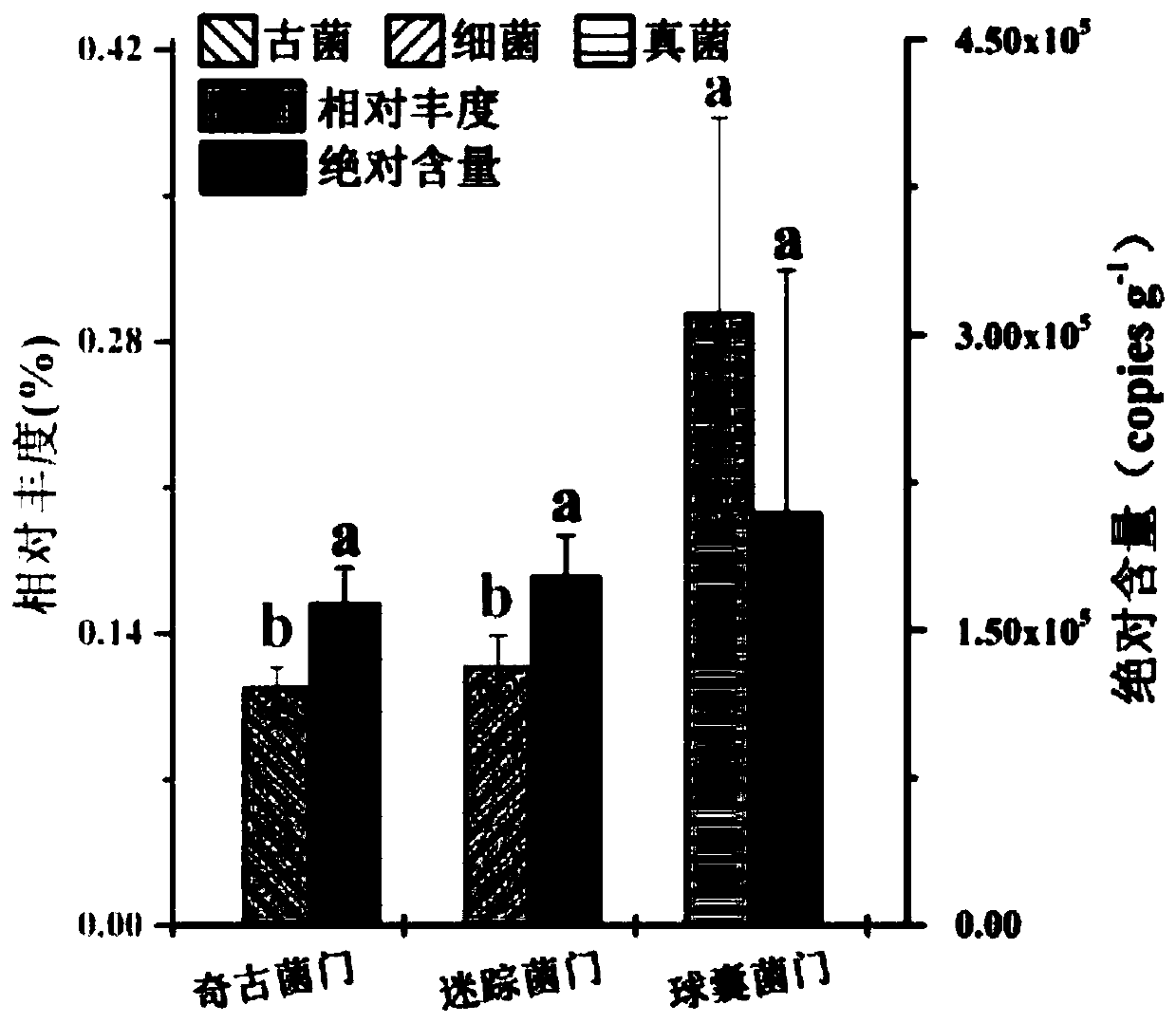
[0052] Embodiment 1. Realize cross-domain comparison in the same soil sample through the three-domain universal internal standard strain, and perform the following steps in sequence:
[0053] (1) Take out the three-domain universal internal standard strain E.coli WYL stored at -80°C, pipette 200μL into 20mL sterile liquid LB medium, and culture at 37°C and 250rpm for 14-16h.
[0054] (2) Transfer the bacterial liquid obtained in step (1) to a sterile 50 mL centrifuge tube, centrifuge at 6000×g for 5 min to obtain bacterial cells, and discard the supernatant.
[0055] (3) First add 5 mL of 4°C pre-cooled sterile water to the centrifuge tube, resuspend the bacteria, then add 25 mL of sterile water and mix well.
[0056] (4) Place the centrifuge tube in a high-speed centrifuge, centrifuge at 6000×g for 5 minutes to obtain bacteria, and discard the supernatant.
[0057] (5) Repeat steps (3)-(4) twice.
[0058] (6) Resuspend the obtained bacteria in sterile water, and adjust the ...
Embodiment 2
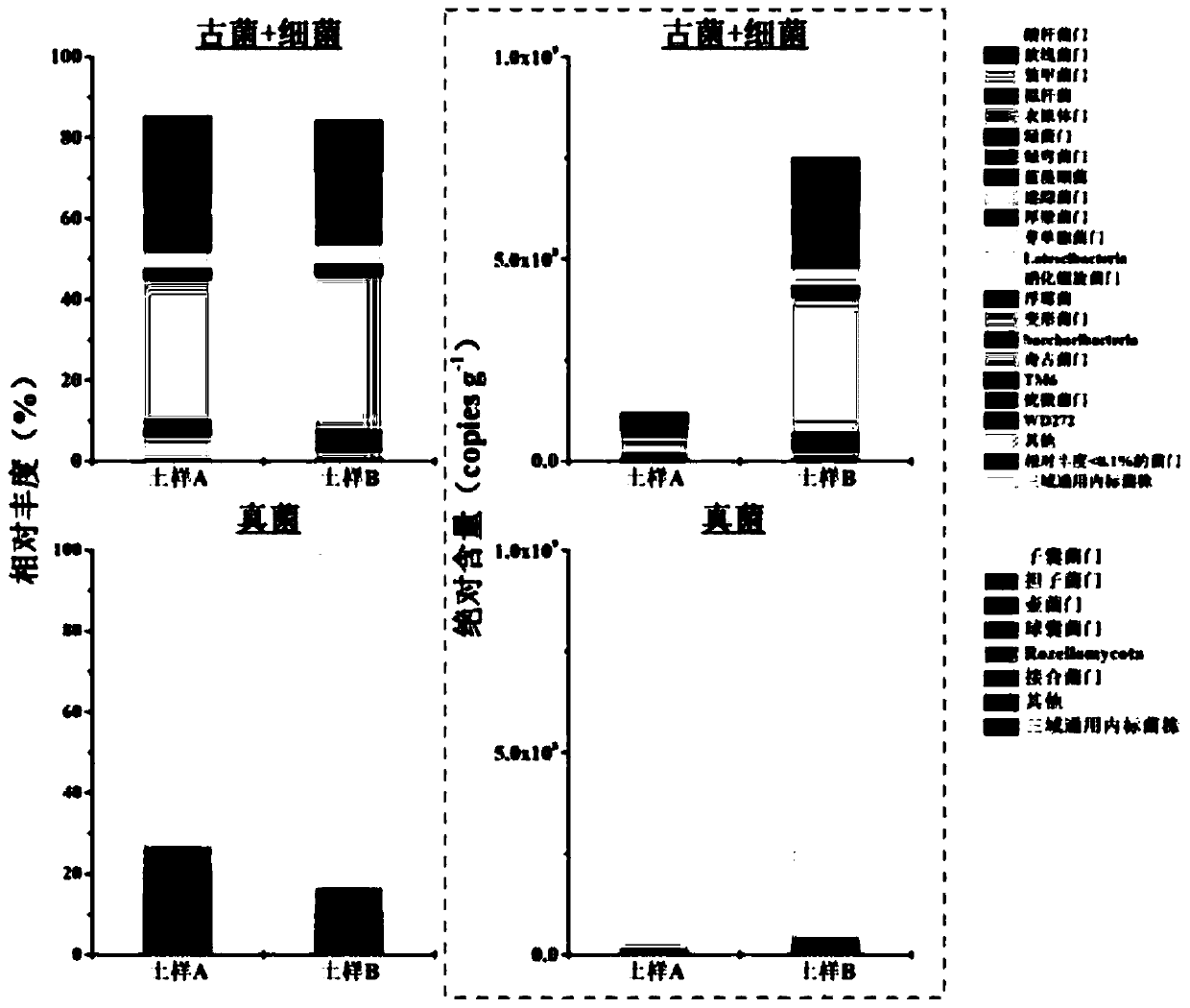
[0082] Example 2. Cross-sample and cross-domain comparison of two soil samples through the three-domain universal internal standard strain
[0083] (1) Take out the three-domain universal internal standard strain E.coli WYL stored at -80°C, pipette 200μL into 20mL sterile liquid LB medium, and culture at 37°C and 250rpm for 14-16h.
[0084] (2) Transfer the bacterial suspension in (1) to a sterile 50mL centrifuge tube, centrifuge at 6000×g for 5min to obtain bacterial cells, and discard the supernatant.
[0085] (3) First add 5 mL of 4°C pre-cooled sterile water to the centrifuge tube, resuspend the bacteria, then add 25 mL of sterile water and mix well.
[0086] (4) Place the centrifuge tube in a high-speed centrifuge, centrifuge at 6000×g for 5 minutes to obtain bacteria, and discard the supernatant.
[0087] (5) Repeat steps (3)-(4) twice.
[0088] (6) Resuspend the obtained bacteria in sterile water, and adjust the absorbance OD of the bacteria suspension 600nm to aroun...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com