Protein and micromolecule binding site predicting method and predicting device
A technology combining sites and prediction methods, applied in proteomics, ICT adaptation, genomics, etc., can solve the problem of limited prediction effect, and achieve the effect of good characterization effect and good prediction effect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0047] The present invention will be further described in detail below in conjunction with specific embodiments.
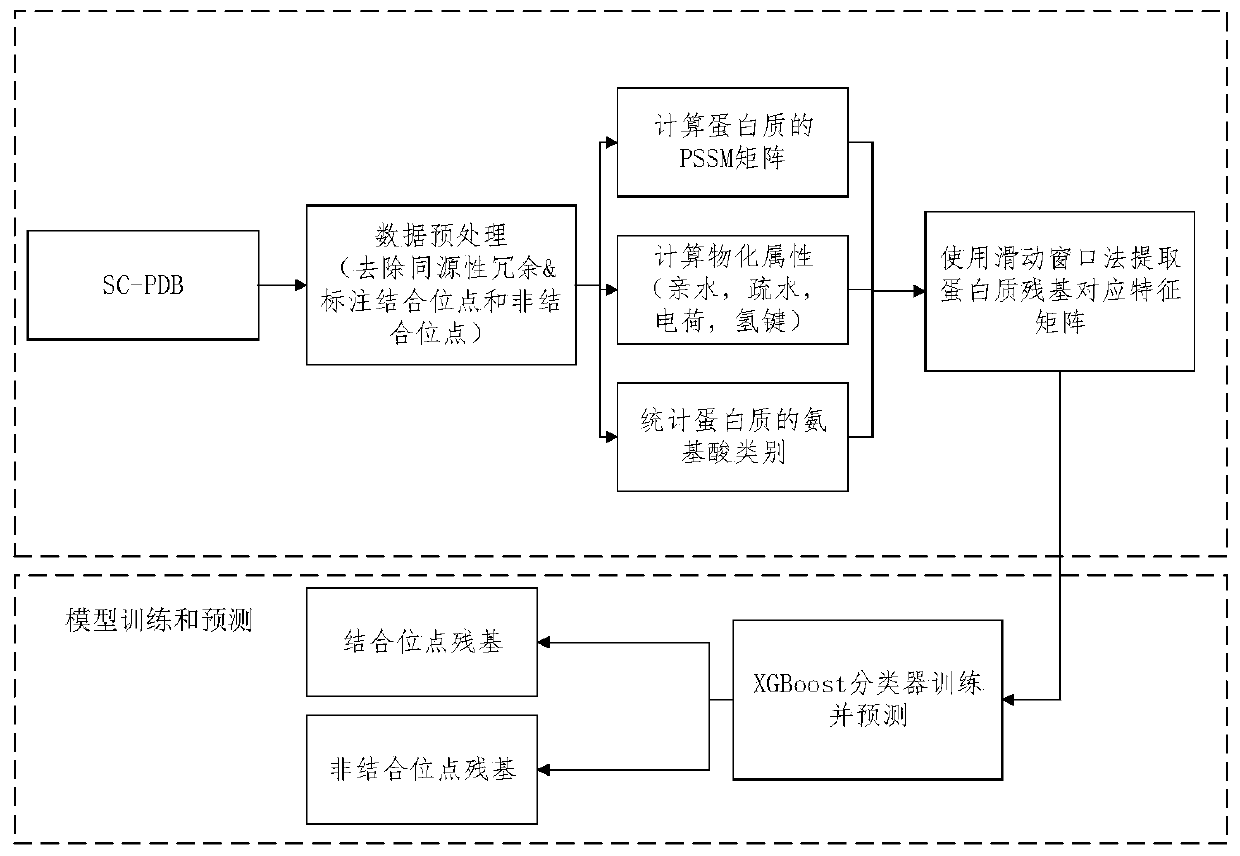
[0048] Example 1 of the method for predicting binding sites of proteins and small molecules
[0049] In the present invention, a new method for predicting the binding sites of small molecules and proteins based on the PDB data of protein small molecules is proposed. The classification model constructed in the experiments of the present invention can be used to predict binding sites and non-binding sites. By extracting features such as the physicochemical properties and evolution information of residues on the protein sequence, generate the data sets required for training and testing, train and test the classification model, and finally use the classification model to classify and predict the binding sites and non-binding sites of proteins point. Protein PSSM matrix information has been used by many scientists, such as predicting binding sites, secondary structure...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com