Development method and application of saccharum cultivar genome SSR (simple sequence repeat) molecular marker
A technology of molecular markers and cultivars, applied in biochemical equipment and methods, recombinant DNA technology, DNA/RNA fragments, etc., can solve the problem of backward construction of genetic linkage map of sugarcane SSR molecular markers, limited number of SSR molecular markers, and great difficulty in development To achieve the effect of improving the level of sugarcane genetics and breeding, enriching polymorphism information, and protecting legitimate rights and interests
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] A total of 4,460 sequences were downloaded from the Sugarcane Genome Center of the French Agricultural Research Institute (http: / / sugarcane-genome.cirad.fr) using the above method, 27,241 SSR sites were found, and 22,932 pairs of SSR primers were successfully designed. The specific operations are as follows :
[0023] From 22,932 pairs of designed primers, search for SSR primers with TG and CA motif types, randomly select 50 pairs, and use 4 Saccharum materials (cultivar R570, cultivar ROC1, tropical species LA purple and cut hands) SES 208) for amplification efficiency verification (see Table 1). The amplification results showed that: a total of 45 pairs of primers could amplify clear amplification bands, the remaining 5 pairs of primers had no amplification bands or the amount of amplified products was weak, and another 35 pairs of primers showed multiple The polymorphic rate was 70% (35 / 50), among which there were 28 pairs of primers of TG repeat type and 7 pairs of p...
Embodiment 2
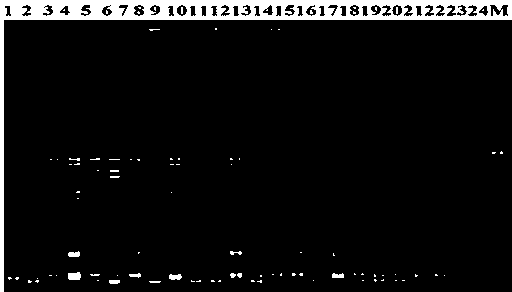
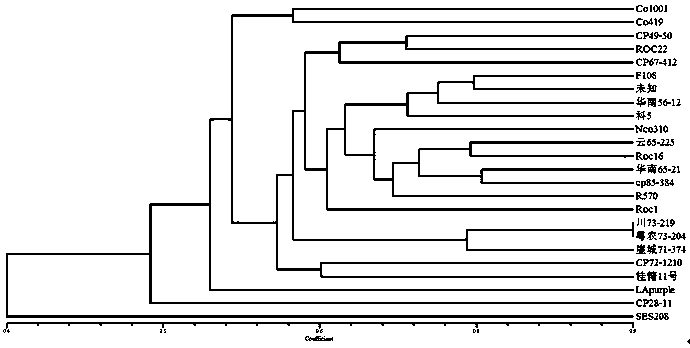
[0027] In order to further verify the polymorphism of the SSR primer pairs identified in this study, 20 pairs of SSR primers with high polymorphism were selected from the 35 pairs of primers screened above, and the 18 backbone parents in my country (whose blood relationship comes from Tropical species, 2-4 species of sarcophagus, large-stem wild species and Indian species) (see Table 2), 2 sugarcane ancestor species (saccharomyces SES 208 and tropical species LApurple) and 4 worldwide Important sugarcane cultivars (LCP85-384, R570, ROC 16 and ROC 22) were analyzed for genetic diversity and polymorphism evaluation of SSR primers. The results showed that: 20 pairs of primers showed obvious polymorphisms on 24 sugarcane experimental materials, 95 alleles were amplified in total, 1-7 alleles were amplified by each pair, and each pair of primers amplified on average 4.75 alleles were identified. figure 2 The PCR amplification electrophoresis patterns of 2 pairs of SSR primers on 24...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com