A method for identification of mixed pseudo- and anti-estrogen disruptors based on enhanced sampling molecular dynamics simulation
A technology of molecular dynamics and anti-estrogen, applied in the direction of informatics, computational theoretical chemistry, instruments, etc., can solve limitations and other problems, and achieve the effect of low cost and high efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] Example 1: Molecular dynamics simulation of ERα
[0067] The human ERα structure was obtained from the ERα protein structure with PDB code 3erd obtained from the protein database, and the integrity of the structure was checked and the default residues were repaired by the Swiss-PdbViewer software. Ligand small molecules, including standard substances and endocrine disruptor bisphenols commonly found in the environment, are structurally optimized and docked in the receptor using the Surflex-Dock module in SYBYL 7.3 to form a ligand-receptor complex. The complex is subjected to enhanced sampling molecular dynamics simulation using the widely recognized enhanced sampling molecular dynamics simulation method—metadynamics simulation method—and the molecular simulation software used is gromacs and plumed software packages. The distance between L544 and E380 and between L544 and M522 in ERα is used as the set variable to carry out metadynamics simulation. The time of each meta...
Embodiment 2
[0068] Embodiment 2: analysis of molecular dynamics simulation results
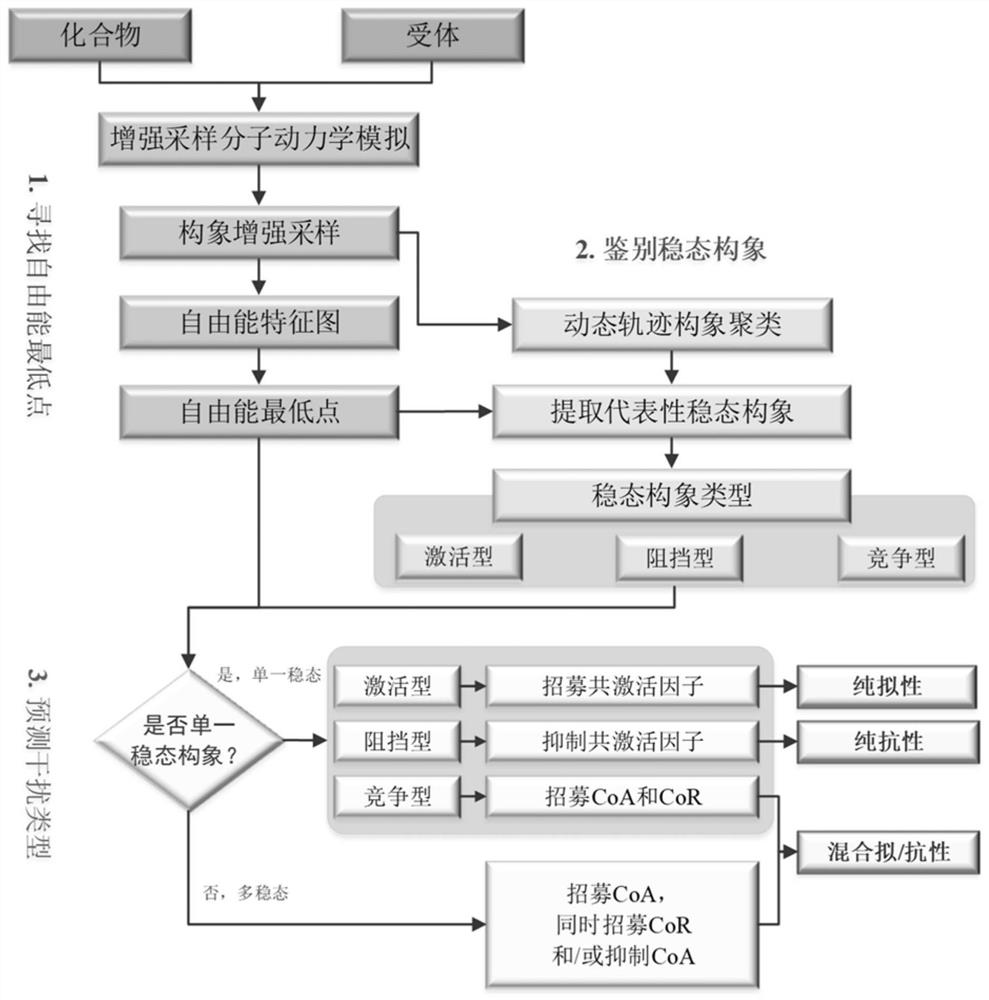
[0069]The molecular simulation trajectory obtained from the metadynamics simulation carried out in the present invention is used for further analysis. The dihedral angle formed by the α-carbon atoms on the four amino acids M543, L539, A350, and L354 of ERα is used as the set variable 1 (CV1, in radians), and the angle formed by the α-carbon atoms on the three amino acids L539, M534, and M522 For CV2 (unit is radian), draw the free energy (Free Energy, unit is kJ / mol) feature map, describe the position of H12, get the lowest point of global and local free energy, and obtain the lowest point of free energy through conformational clustering The representative conformation is the representative steady-state conformation. According to the H12 stable position of the representative steady-state conformation, the type of each steady-state conformation was judged. Finally, according to the type and quantity of t...
Embodiment 3
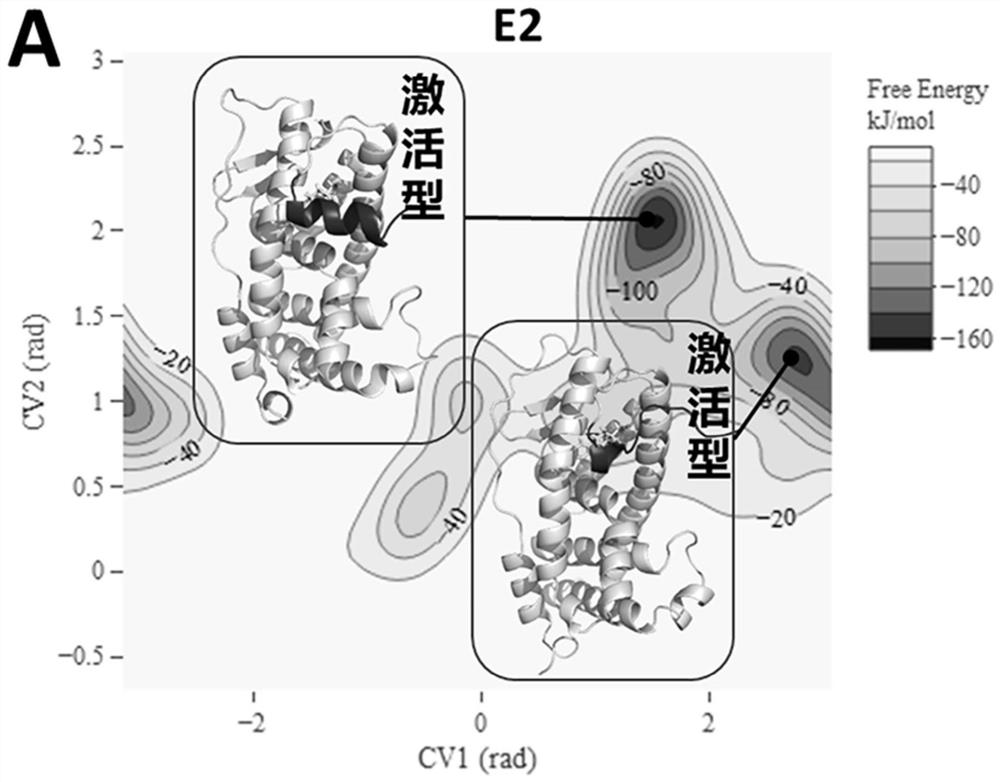
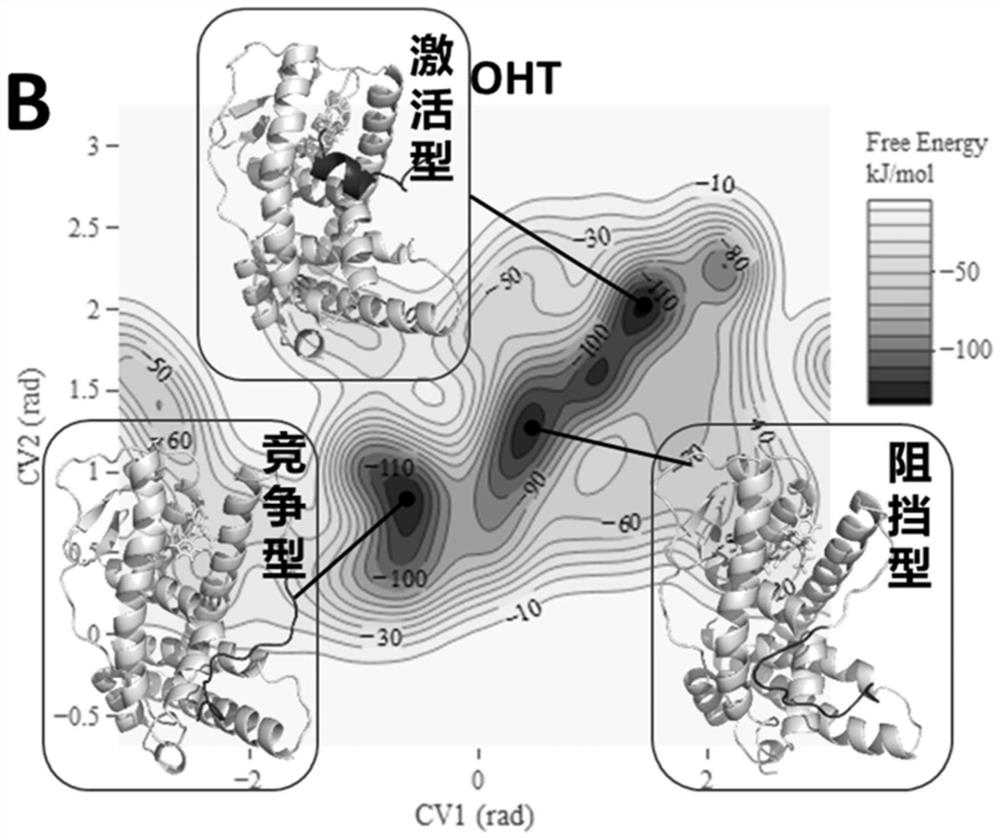
[0070] Embodiment 3: the result analysis of standard substance E2, OHT
[0071] For E2 of the pure pseudo-standard substance, the metadynamics simulation results show that the steady-state conformation of E2-ERα is the active conformation ( Figure 2A ), which is consistent with the reported crystal structure of E2-ERα. The activated conformation of H12 forms a "steric hindrance" effect to selectively recruit CoA. Therefore, under the action of pure mimetic compound E2, ERα forms an activated conformation, leading to the selective recruitment of CoA by the receptor, which in turn leads to transcriptional activation and pure mimetic effects. For the OHT of mixed pseudo-resistant compounds, the metadynamics simulation results show that OHT-ERα has various types of steady-state conformations ( Figure 2B ). OHT-ERα has three types of steady-state conformations: active, blocking and competing, and the blocking and competing conformations are consistent with the reported crystal...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com