Method to generate biocompatible dendritic polymers for analyte detection with multimodal labeling and signal amplification
A technology for polymers and analytes, used in biochemical equipment and methods, determination/inspection of microorganisms, analytical materials, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] molecular mechanism
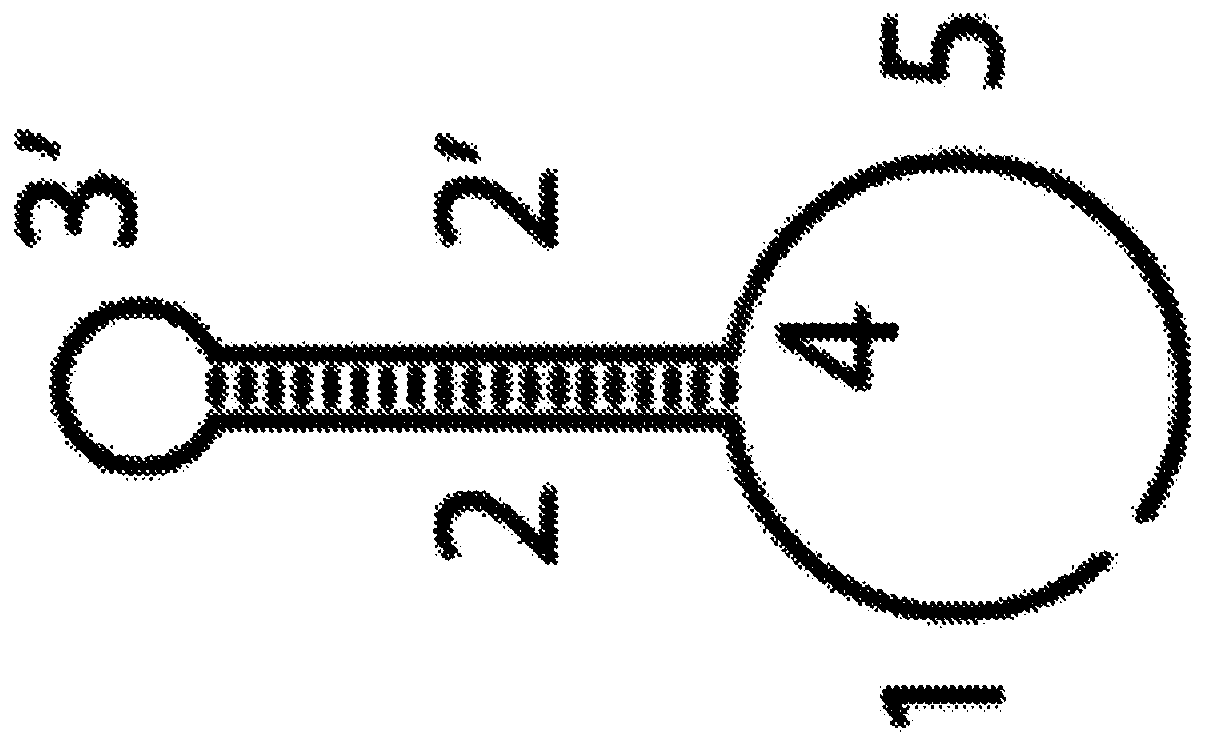
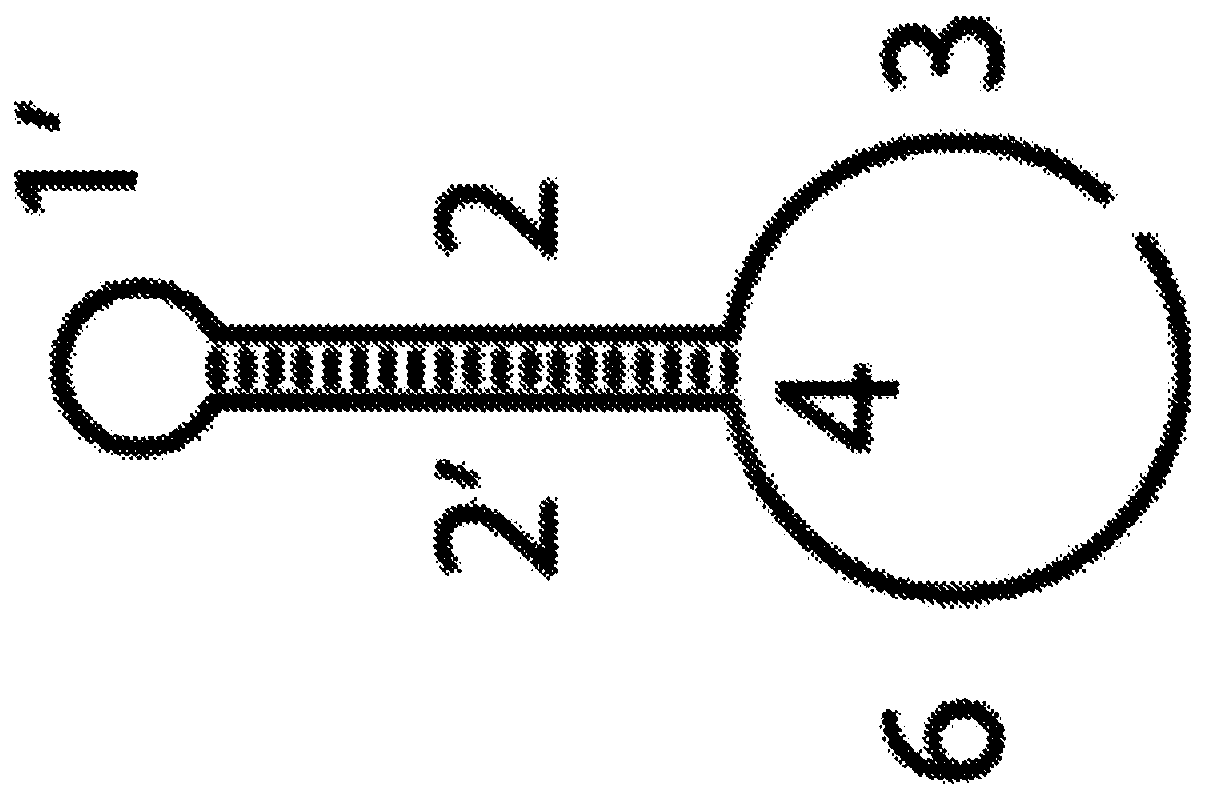
[0078] The inventors' method works in the following way: Complementary pairs of dendrimers can be used to generate dendrimers by self-assembly in a controlled manner via a chain reaction of branch migration and nucleic acid hybridization in the presence of a polymerization trigger objects (Figure 1 and figure 2 ). Triggers can be used directly or in combination with affinity ligands (Figure 3 and Figure 4 ). The nature of the affinity ligand determines the type of analyte to be detected. For example, each branch of the polymer can be used to attach a label of choice, such as a fluorophore, quantum dot, or metal-chelating polymer ( Figure 5 ). Each dendrimer contains about two hundred branches. Consequently, a large number of labels can eventually be attached to the target of interest, greatly multiplying its signal and enabling rapid and unambiguous detection. Following polymerization, labeling can be accomplished by directly hybridizing ...
Embodiment 2
[0080] Advantage
[0081] Another consideration: Certain fluorophores and other signaling labels are incompatible with hairpin oligonucleotides due to secondary structure effects (eg, steric hindrance). Complementary probe labels eliminate these effects and increase synthesis yield relative to direct attachment to long oligonucleotides that can adopt secondary structure. The inventors' approach has several advantages over other approaches capable of achieving similar goals:
[0082] -By generating dendrimers rather than bare directly labeled structures (for example, in HCR), the inventors' method provides greater flexibility and Modularity. This will enable the user to select the most appropriate marker selected for the specific situation. This provides flexibility to the user such that various detection methods that can be used orthogonally to the methods described herein provide complementary information.
[0083] - Compared to HCR, the inventors' method significantly re...
Embodiment 3
[0089] Dendritic Amplifier Design Considerations
[0090] The described method allows an extremely large number of sequence designs for marker combinations. For example, for a system with a foothold / loop of 12 nucleotides in length, a stem of 24 nucleotides in length, and branches of 15 nucleotides in length, there are 4 12 ×4 24 ×4 12 ×4 15 =4 63 =8.5×10 37 possible sequence. This incredibly large design space offers the possibility to generate very large or orthogonal systems. The inventors have established design criteria for producing an optimal scale-up system.
[0091] For example, an ideal sequence combination satisfies certain design criteria including: minimizing alternative conformations of preferred hairpin secondary structures, minimizing interactions between footholds and branches, and maximizing stem stability. In more detail, sequences are selected to stack bases in key positions (e.g., the leading edge of the foothold, the last position of the foothold ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com