Device for detecting blood disease fusion gene
A fusion gene and blood disease technology, applied in the field of biology, can solve problems such as instability, undetectable fusion type, difficulty in sample storage and transportation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Sample: Known sample, clinically detected positive for BCR-ABL1 fusion.
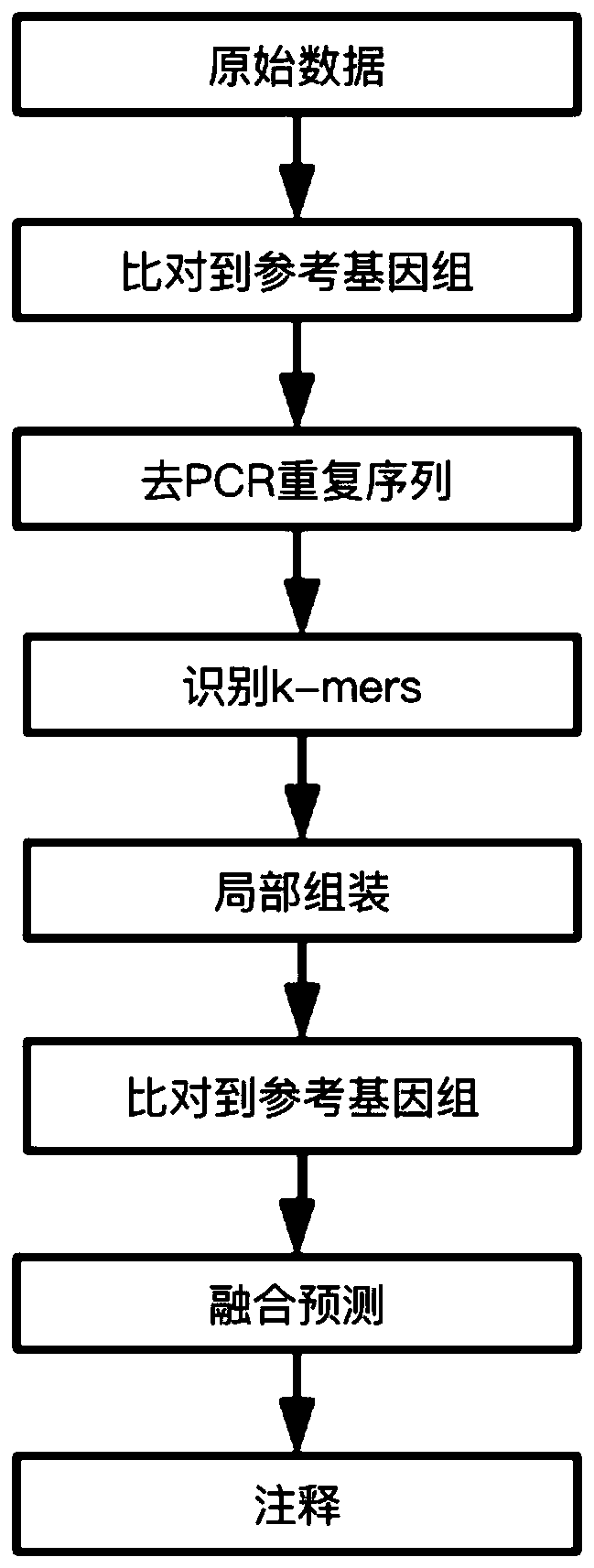
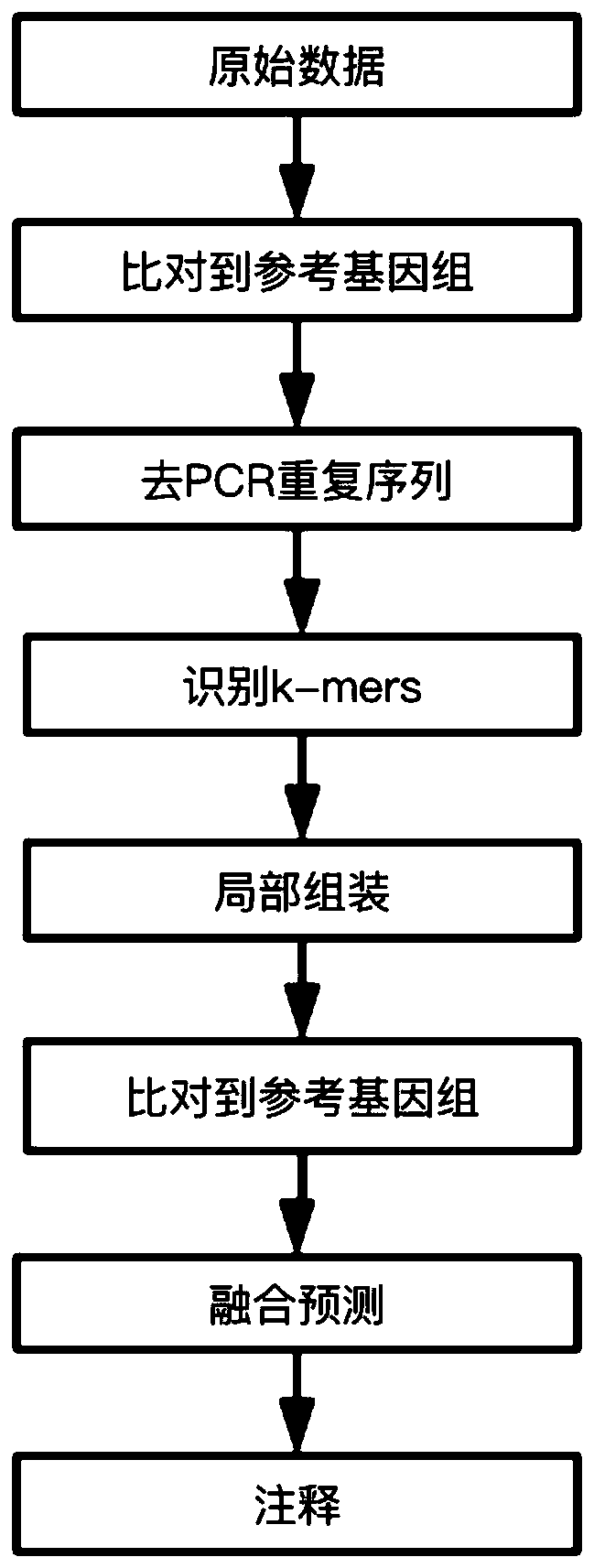
[0049] Use the device of the present invention (execution command reference figure 1 shown):
[0050] 1. The input data is the raw data of next-generation sequencing off-machine, and the data format is fastq.
[0051] 1) Preprocess the original off-machine data, including removing adapters and low-quality data (sequences with N content > 5 or bases below Q15 in the sequence > 40%).
[0052] 2) Compare and sort the processed original off-machine data with the reference genome, and obtain the comparison results, and the data format is bam.
[0053] 3) Identify the duplication reads of the bam file and remove the duplication reads.
[0054] 2. Identify candidate k-mers that may contain breakpoints on the processed alignment data.
[0055] Carry out k-mers recognition in the way of pattern growth, assuming a sequence M of length N is given, k-mer refers to a sequence of length k, this sequence is ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com