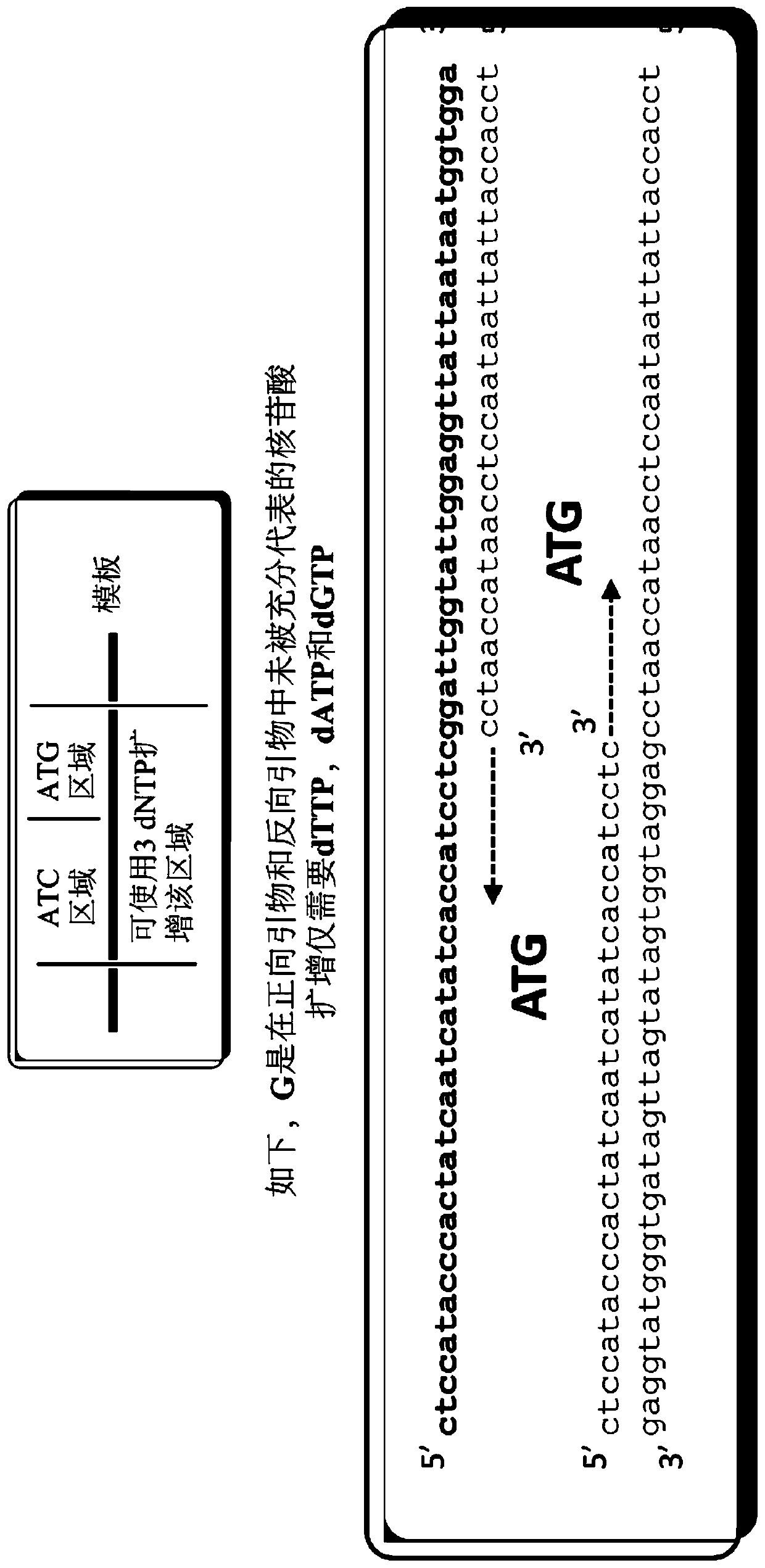
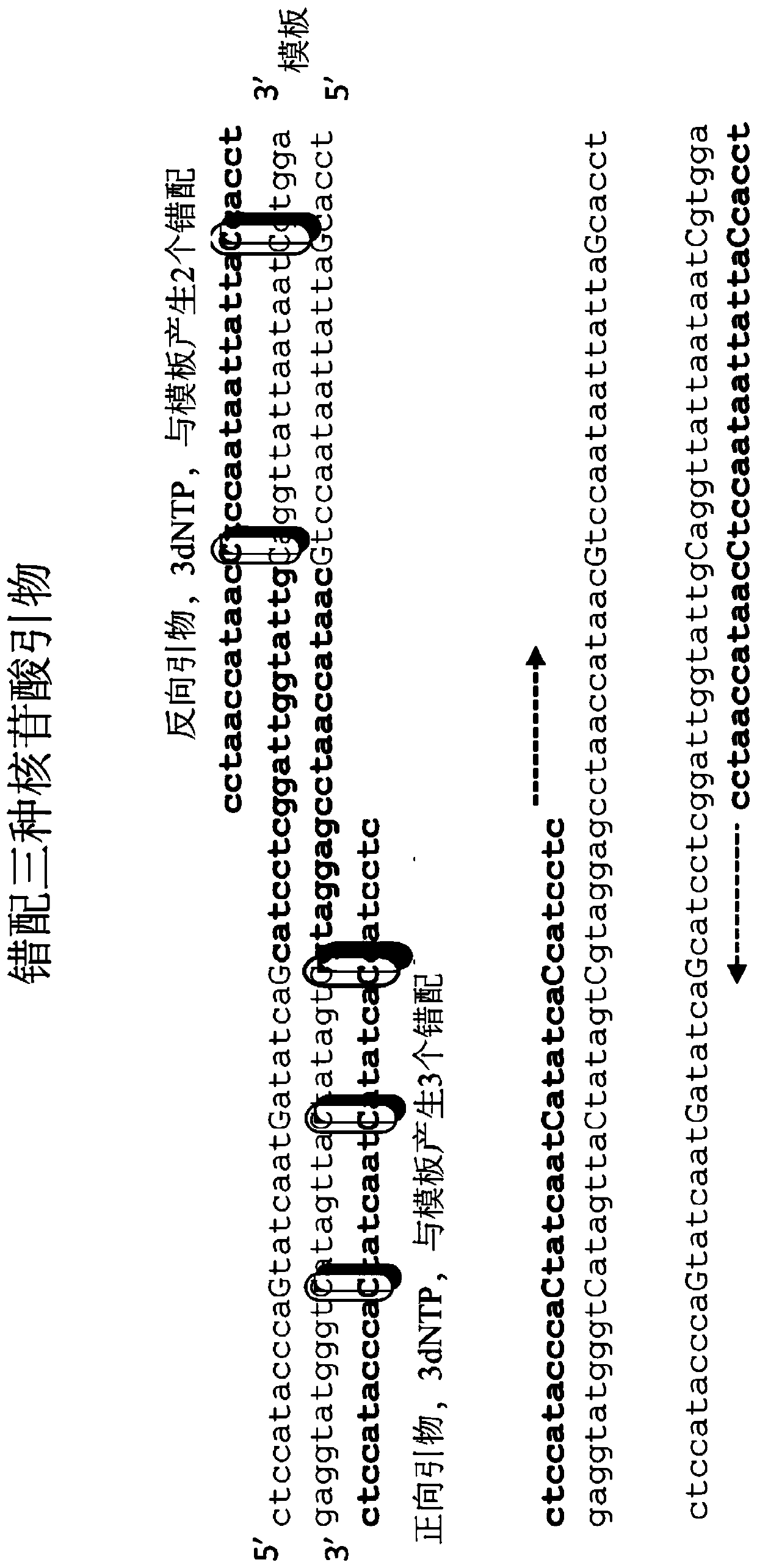
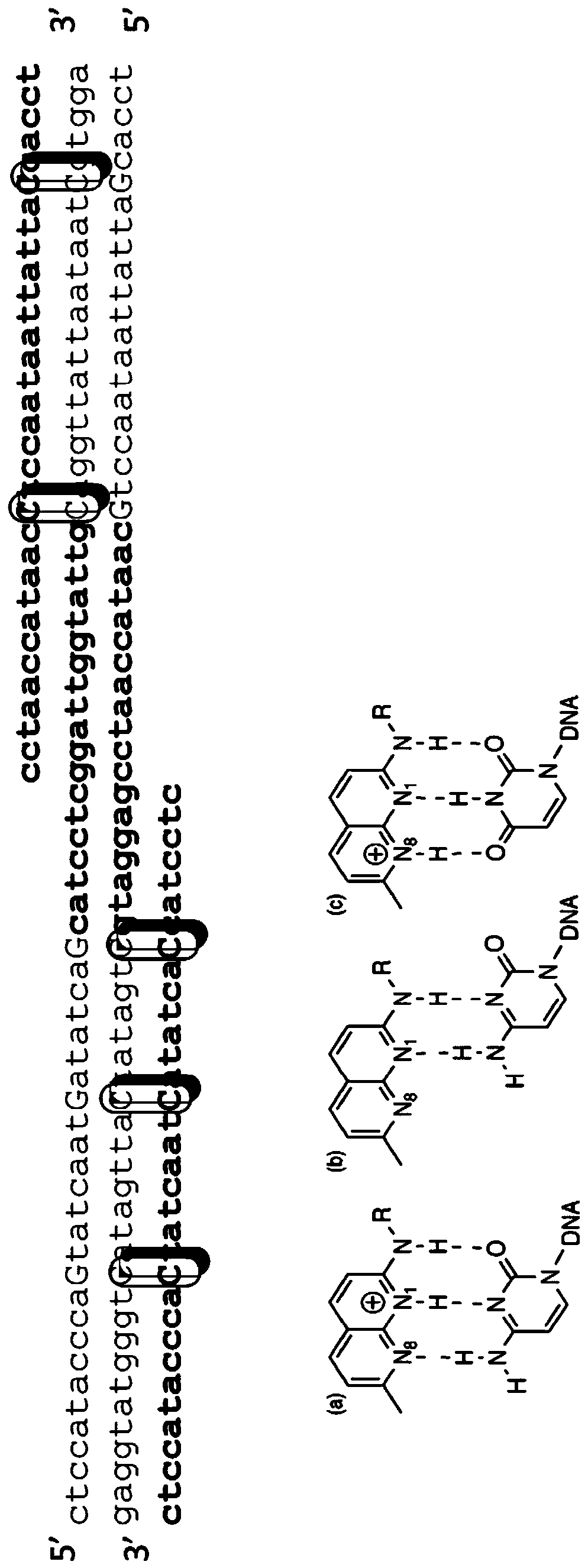
Digital amplification with primers of limited nucleotide composition
A digital amplification and nucleic acid technology, which is applied in the determination/testing of microorganisms, biochemical equipment and methods, fermentation, etc., and can solve complex problems such as interpretation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0238] Example 1: Underrepresented primers reduce background signal in dPCR
[0239] This experiment was performed to determine whether primers with underrepresented nucleotide types (e.g., primers with three nucleotide types) could reduce the dPCR reaction compared to conventional primers containing four nucleotide types. background signal. Primers with underrepresented nucleotides are sometimes referred to as underrepresented primers.
[0240] In this experiment, after performing droplet digital PCR reactions (ddPCR) on template (n=4) and without template (n=2), fluorescence from dPCR reactions containing underrepresented primers was compared to Signals were compared to PCR reactions containing a pair of tetranucleotide primers.
[0241] Briefly, a 20 μL ddPCR reaction containing 1000 copies of the E. coli genomic DNA template, 10 μL QX200 Digital PCR Supermix (2X), and 800 nM of each primer. Primers of the three nucleotide types and conventional primers were made to ...
Embodiment 2
[0245] Example 2: Discrimination of trisomy and euploid DNA
[0246] This experiment was performed to determine whether underrepresented primers could be multiplexed accurately and sensitively in the dPCR platform.
[0247] In this experiment, a 5-multiplex ddPCR reaction was performed in a single reaction tube using five pairs of primers to quantify copy number alterations of chromosome (Chr) 21, chromosome 18, chromosome 13, chromosome X, and chromosome Y.
[0248] 20 μL ddPCR reaction containing 250 copies of template DNA (first or second DNA template type), 10 μL QX200 Digital PCR Supermix (2X), a pair of Chr21 primers, a pair of Chr18 primers, a pair of Chr13 primers, a pair of ChrX primers and a pair of ChrY primers. PCR conditions were as follows: 95°C for 5 minutes, followed by 40 cycles of 95°C for 15 seconds, 60°C for 30 seconds, 4°C for 5 minutes, and heating at 95°C for 5 minutes.
[0249] On average, 11% to 13.4% of the cell-free DNA in maternal blood is of f...
Embodiment 3
[0261] Example 3: Determination of Chromosomal Aneuploidy in cWDNA Using 14-Multiplex dPCR
[0262] In addition to the low abundance of fetal DNA in maternal blood, fetal DNA is also fragmented and enters the bloodstream. Therefore, in noninvasive prenatal testing (NIPT) applications, more amplicons / target chromosomes are required to detect enough positive droplets.
[0263]This experiment was performed to determine whether 14-multiplex dPCR using underrepresented primers in a single reaction tube can accurately quantify and detect very small amounts of trisomy DNA template.
[0264] 20 μL ddPCR reaction contains 500 copies of template DNA (which includes the first, second or third DNA sample), 10 μL QX200 Digital PCR Supermix (2X), Chr21 primer and Chr18 primer. Because dPCR assays designed with more amplicons per target chromosome require a lower amount of cffDNA input, seven different amplicons were chosen for chromosome 21 (Chr18) and seven different amplicons for chr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com