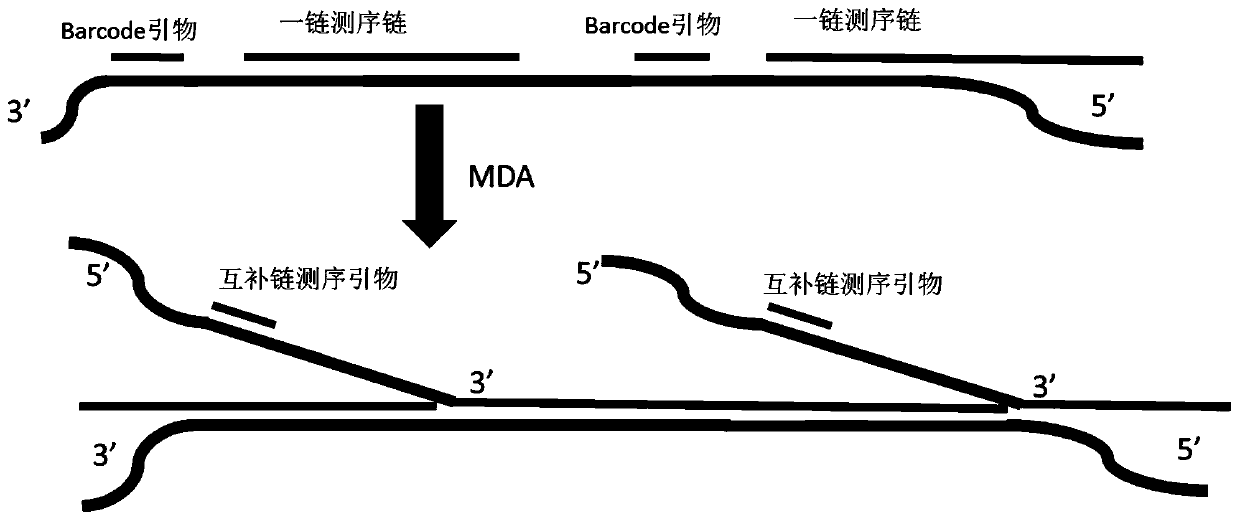
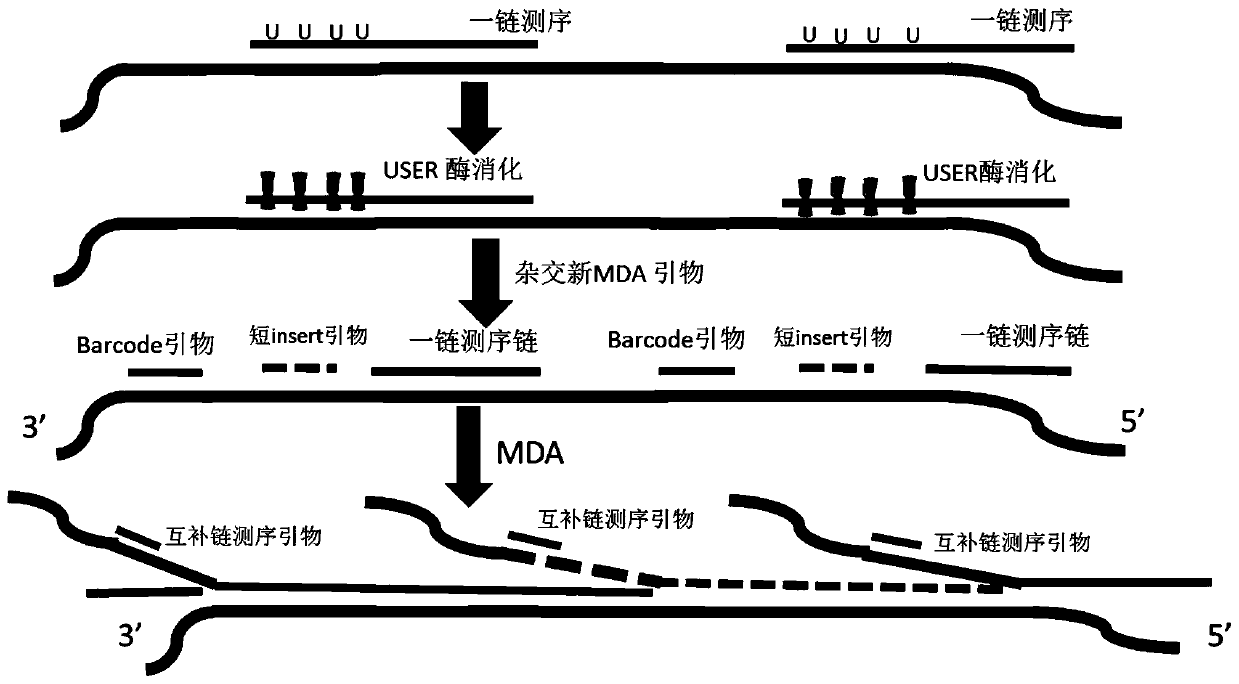
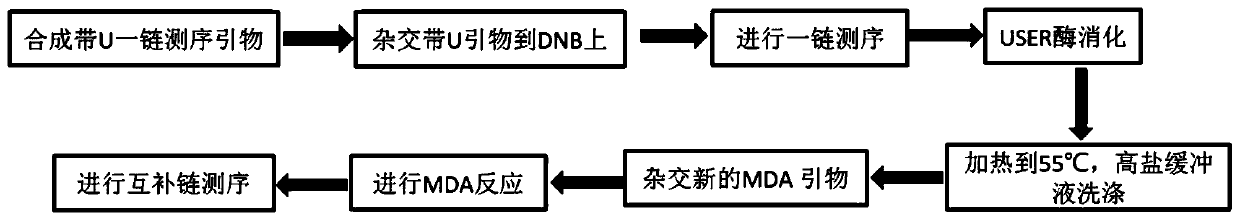
Method for synthesizing DNA nanosphere complementary chains and sequencing method
A technology of nanospheres and complementary chains, applied in the field of sequencing, can solve problems such as difficult hybridization, low sequencing signal, and low fluorescent signal
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0075] Primers and reagents used in the examples:
[0076] Primers for one-strand sequencing:
[0077] CAACTCCUTGGCUCACAGAACGACAUGGCTACGAUCCGACTT (SEQ ID NO: 1);
[0078] MDA Primer 1: AAGTCGGAGGCCAAGCGGTCTTAGAAGACAA (SEQ ID NO: 2);
[0079] MDA Primer 2: CAACTCCTTGGCTCACAGAACGACATGGCT (SEQ ID NO: 3);
[0080] USER enzyme (US NEB company, product number M5505L);
[0081] BGISEQ-500DNB Preparation and Loading Kit (Hua Da Gene Company, Cat. No. 85-05531-00);
[0082] BGISEQ-500 Sequencing Kit (PE100) (BGI Corporation);
[0083] BGISEQ-500 chip (BGI Corporation).
[0084] Taking the BGISEQ-500 platform as an example, the PE100 sequencing effect of the present invention is demonstrated:
[0085] (1) Synthesize a one-strand sequencing primer with U base modification, and dilute it with 5×SSC solution to a working solution concentration of 1 μM. The system is shown in Table 1 below:
[0086] Table 1
[0087]
[0088]
[0089] (2) DNB was prepared using BGISEQ-500DNB Prep...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com