A Genome Annotation Method Using Second- and Third-Generation Transcriptome Sequencing Data
A technology of transcriptome sequencing and genome, applied in genomics, proteomics, instruments, etc., can solve problems such as slow speed, inability to use at the same time, and insufficient accuracy and efficiency of genome prediction, so as to achieve strong operability and improve accuracy , the effect of improving efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0019] The present invention will be further described below with reference to the accompanying drawings and specific embodiments.
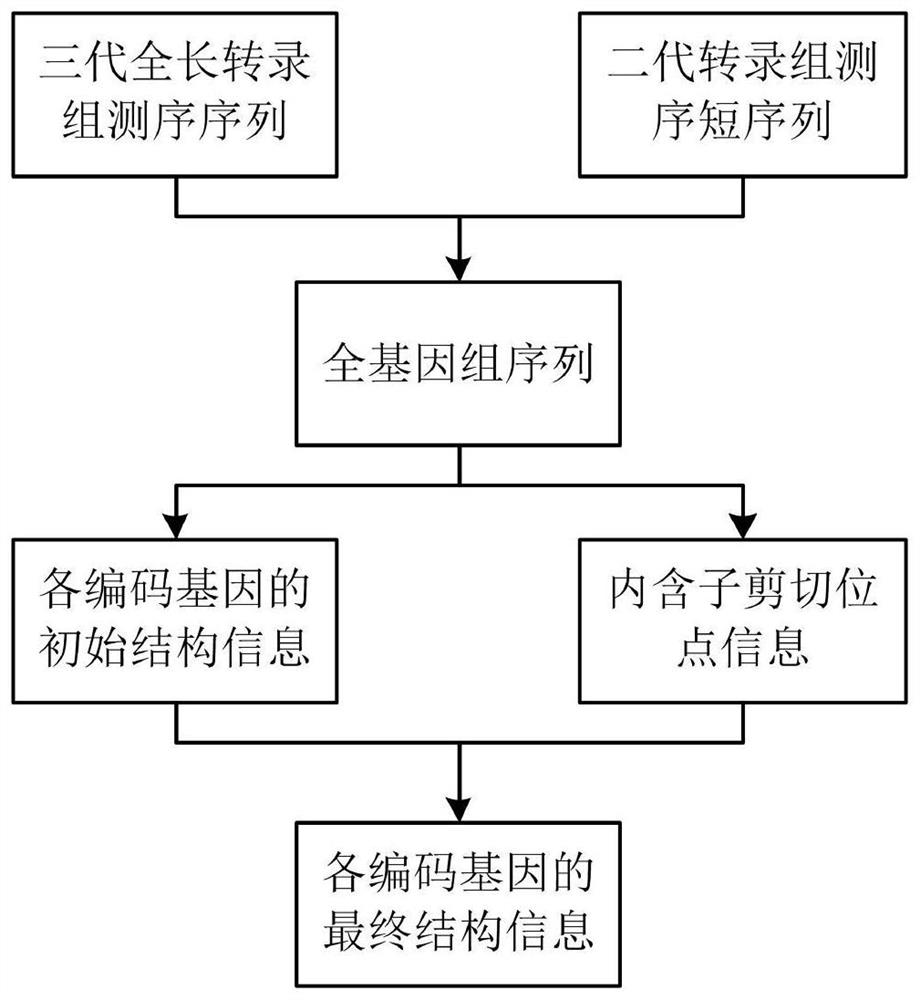
[0020] In this example, the genome annotation method of the present invention is used to annotate the genome structure of plateau fish. like figure 1 As shown, the genome annotation method using the second-generation and third-generation transcriptome sequencing data of the present invention includes the following steps:
[0021] Step 1: Compare the three-generation full-length transcriptome sequencing sequences to the target genome to obtain the initial structural information of each coding gene, which specifically includes the following steps:
[0022] Step 1.1: Predict the protein-coding sequence of the third-generation full-length transcriptome sequencing sequence: analyze the third-generation full-length transcriptome sequencing sequence to obtain a full-length non-chimeric sequence (FLNC), and predict the possible protein-coding sequence o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com