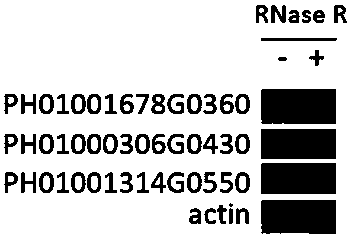
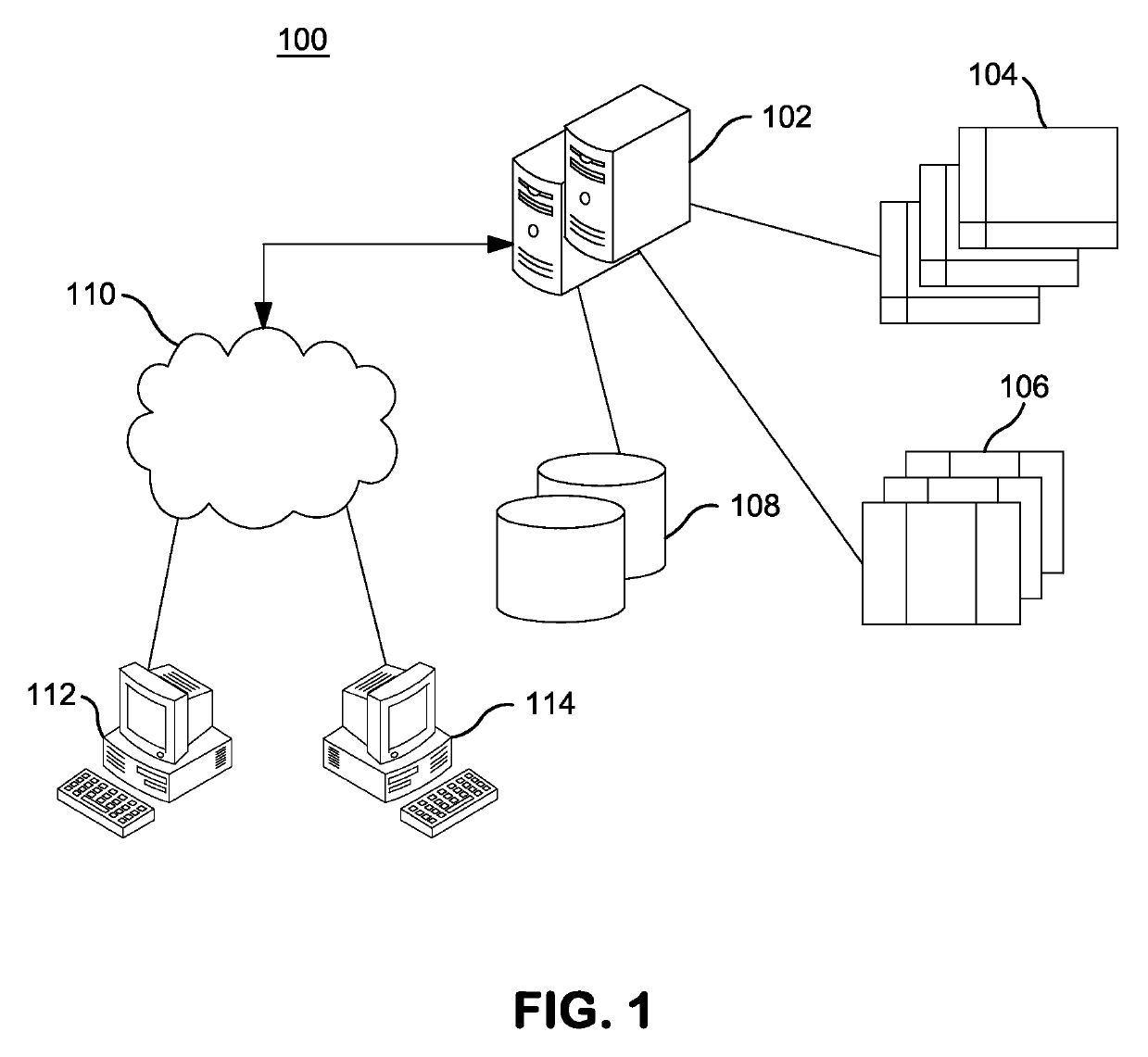
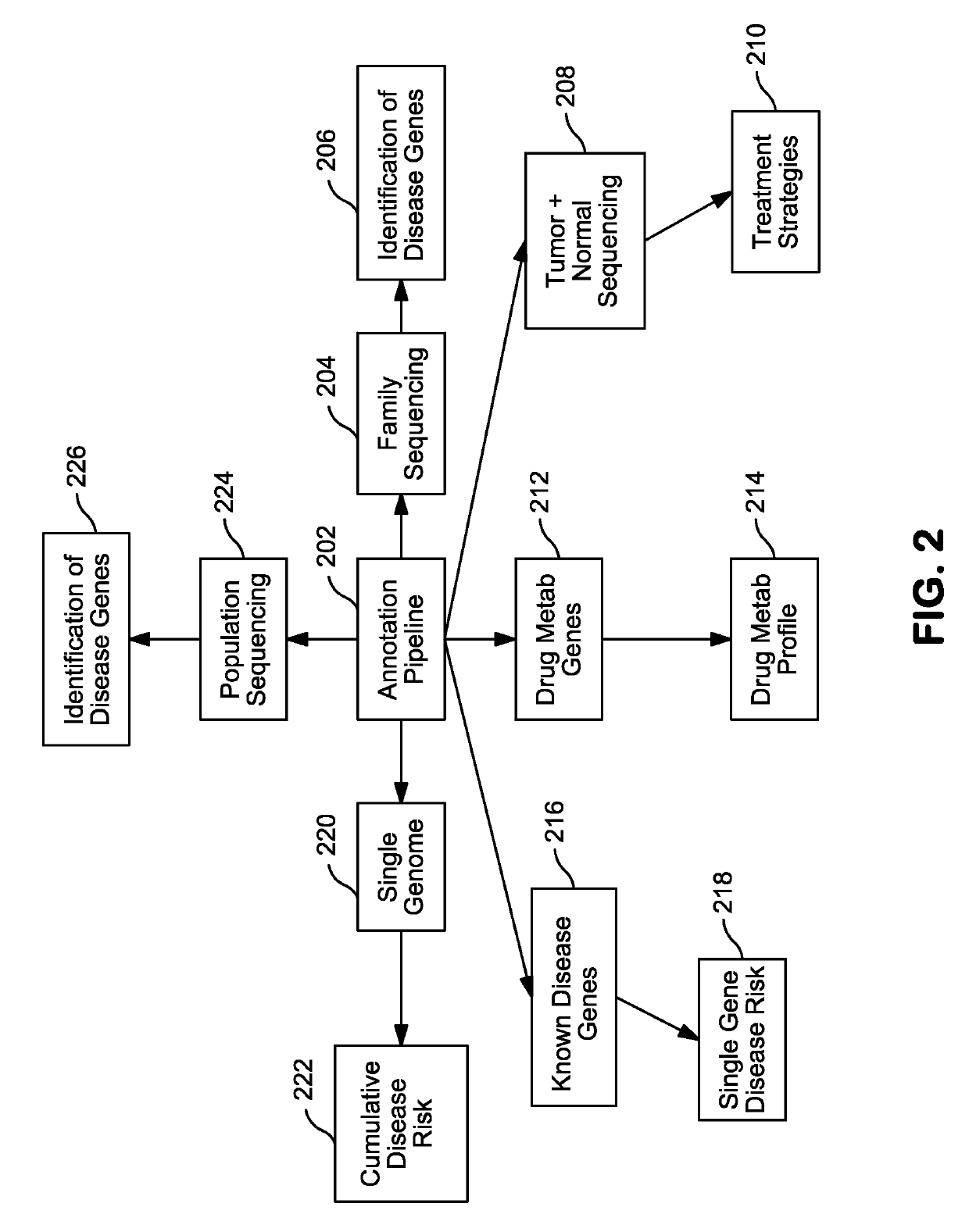
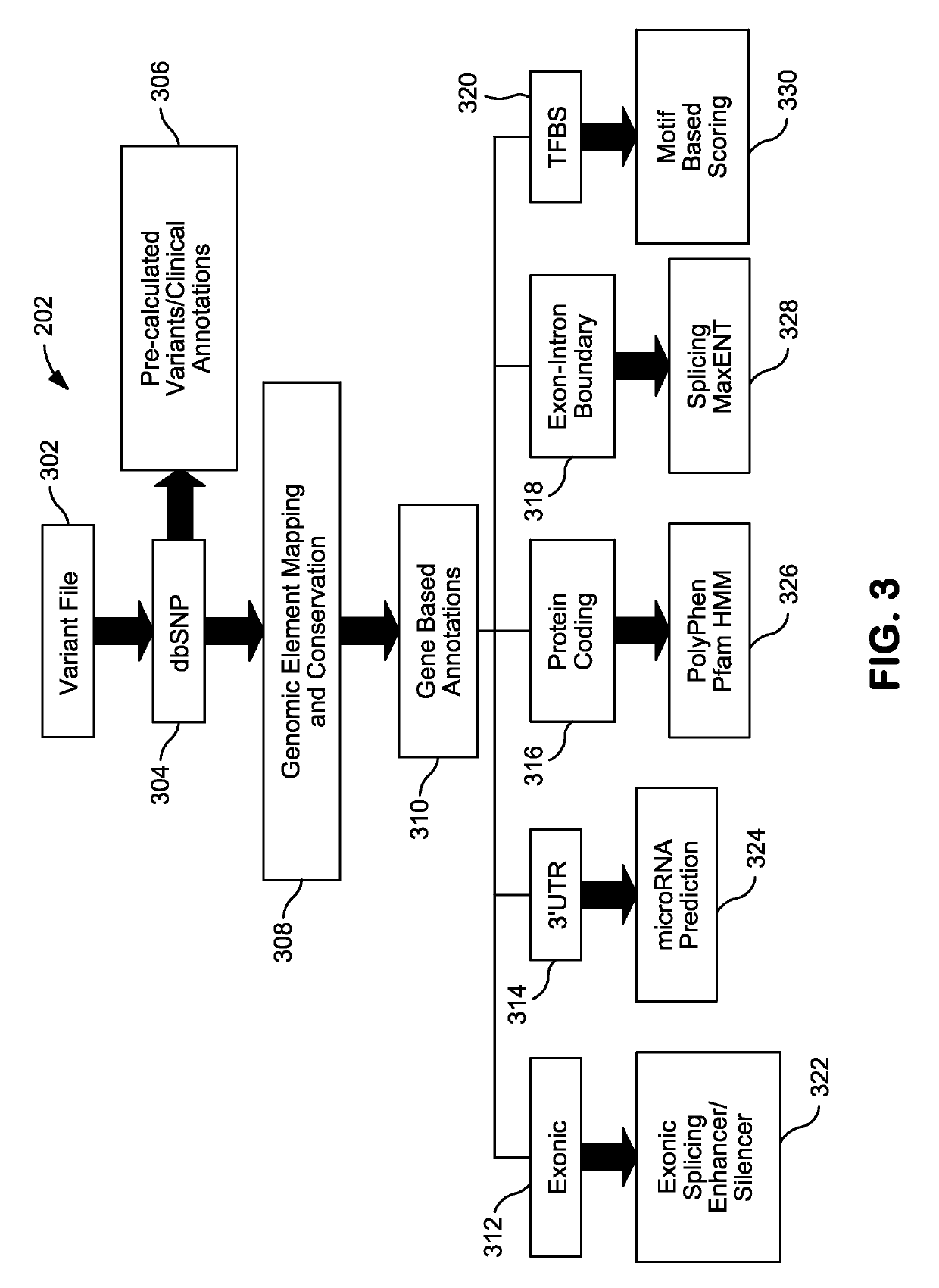
Patents
Literature
31 results about "Genomic annotation" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
DNA annotation or genome annotation is the process of identifying the locations of genes and all of the coding regions in a genome and determining what those genes do. An annotation (irrespective of the context) is a note added by way of explanation or commentary.
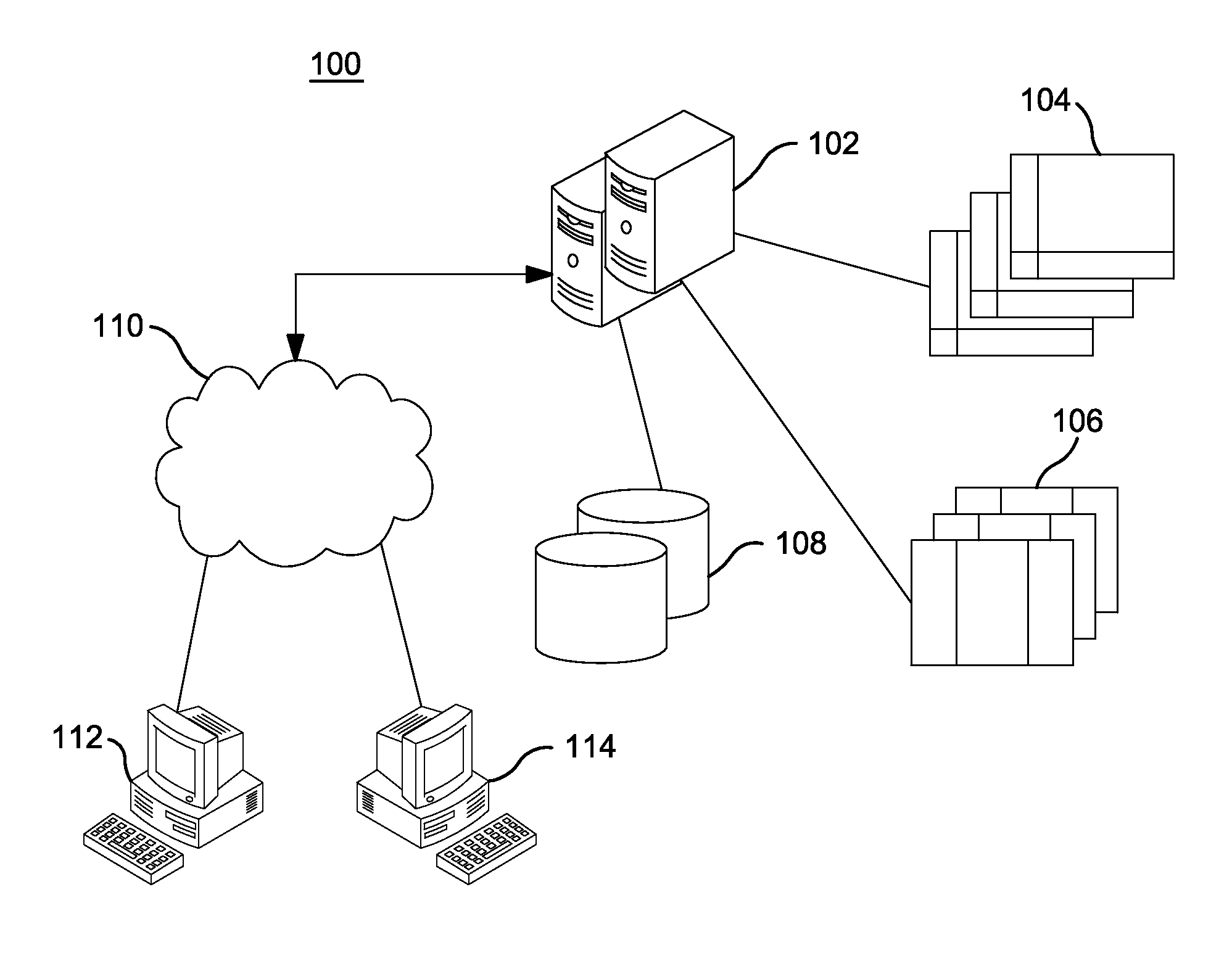
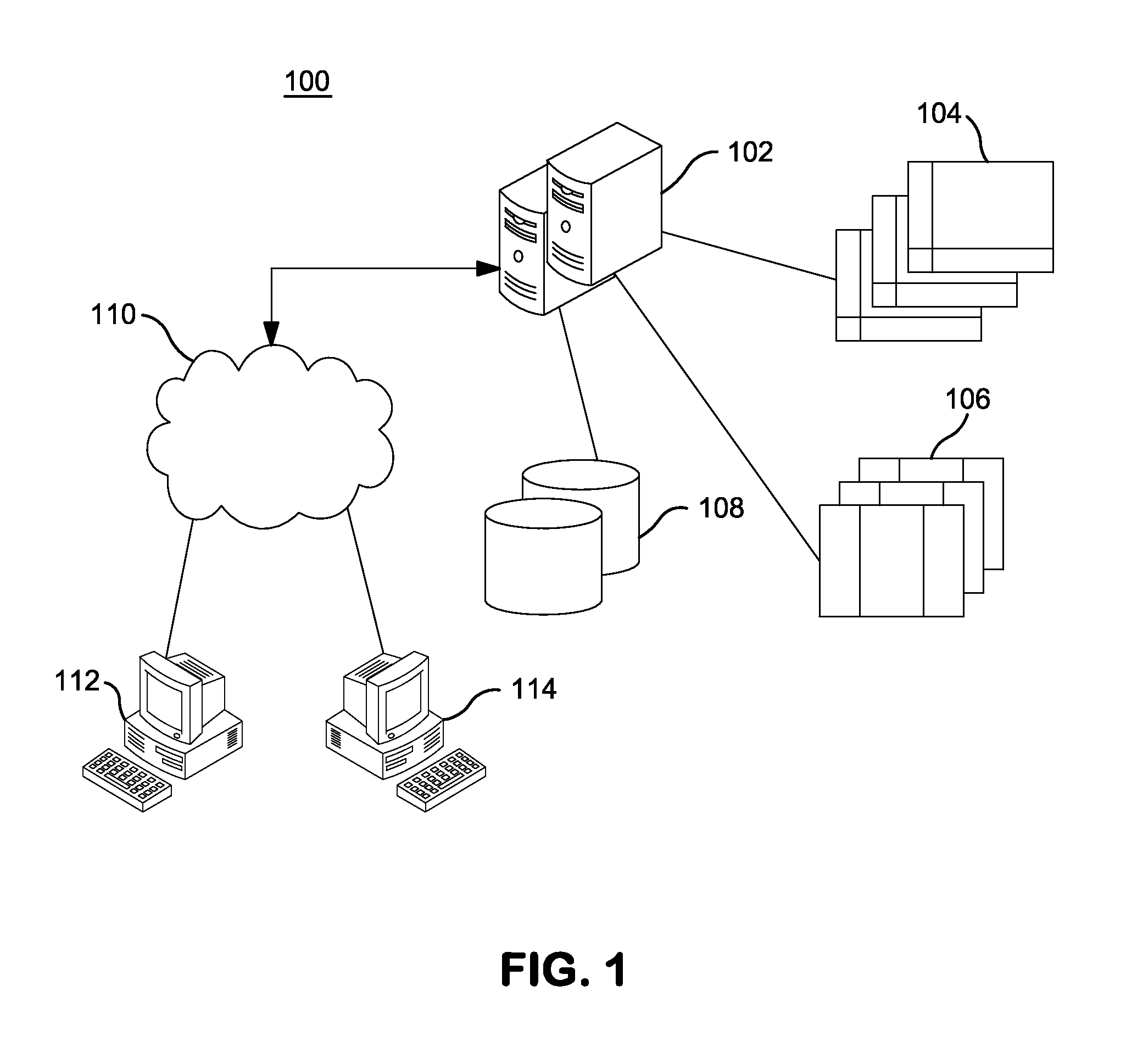
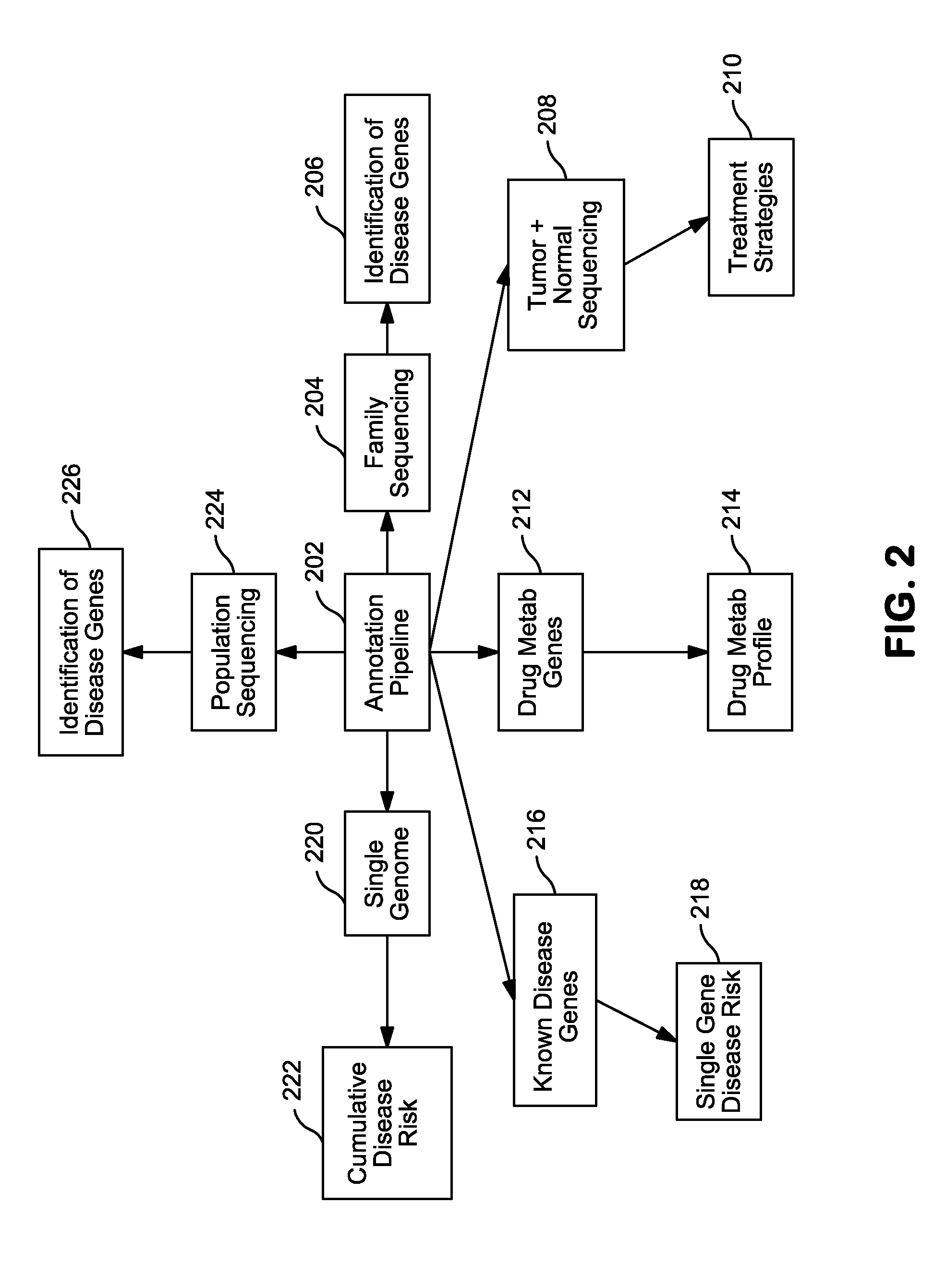
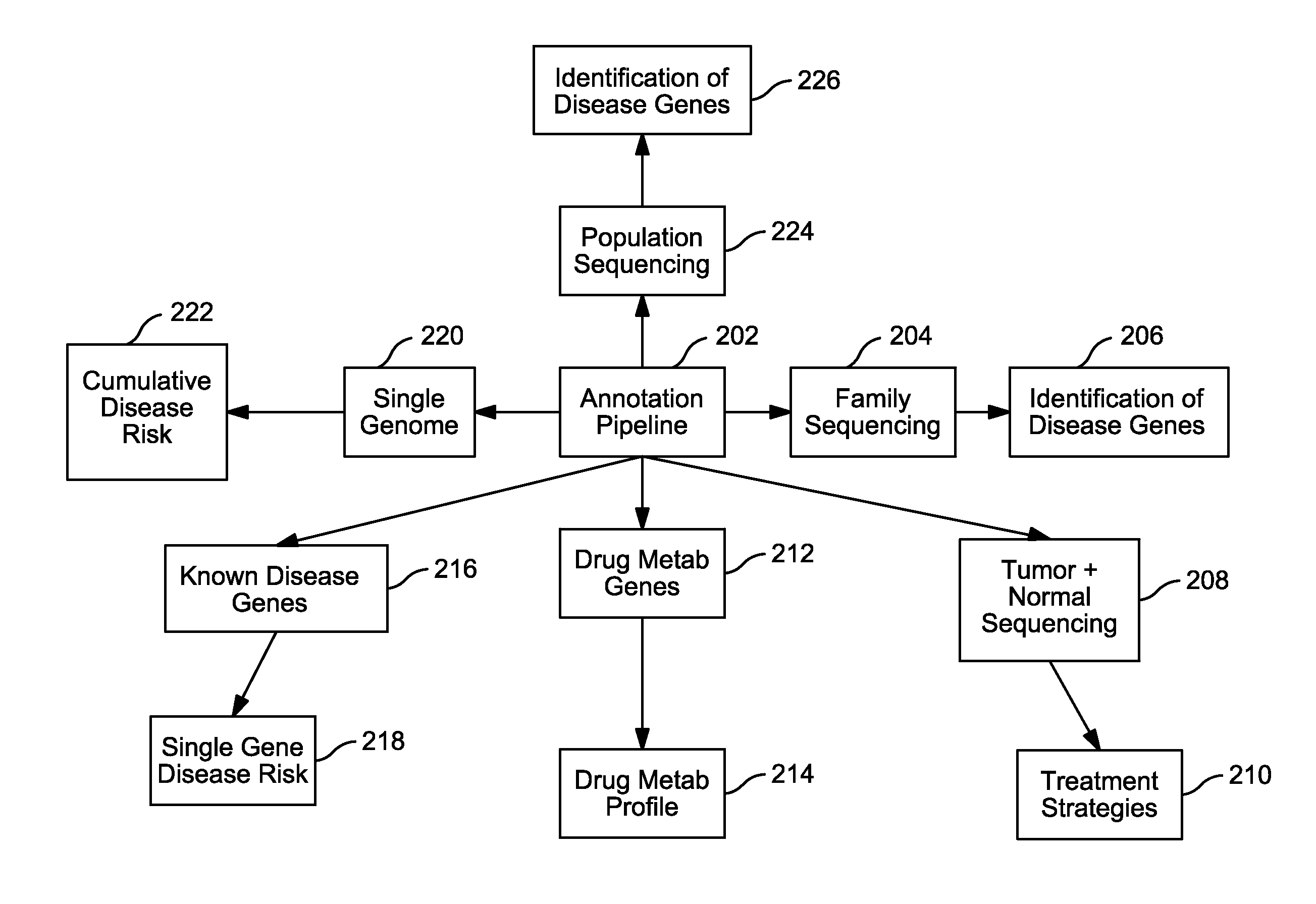
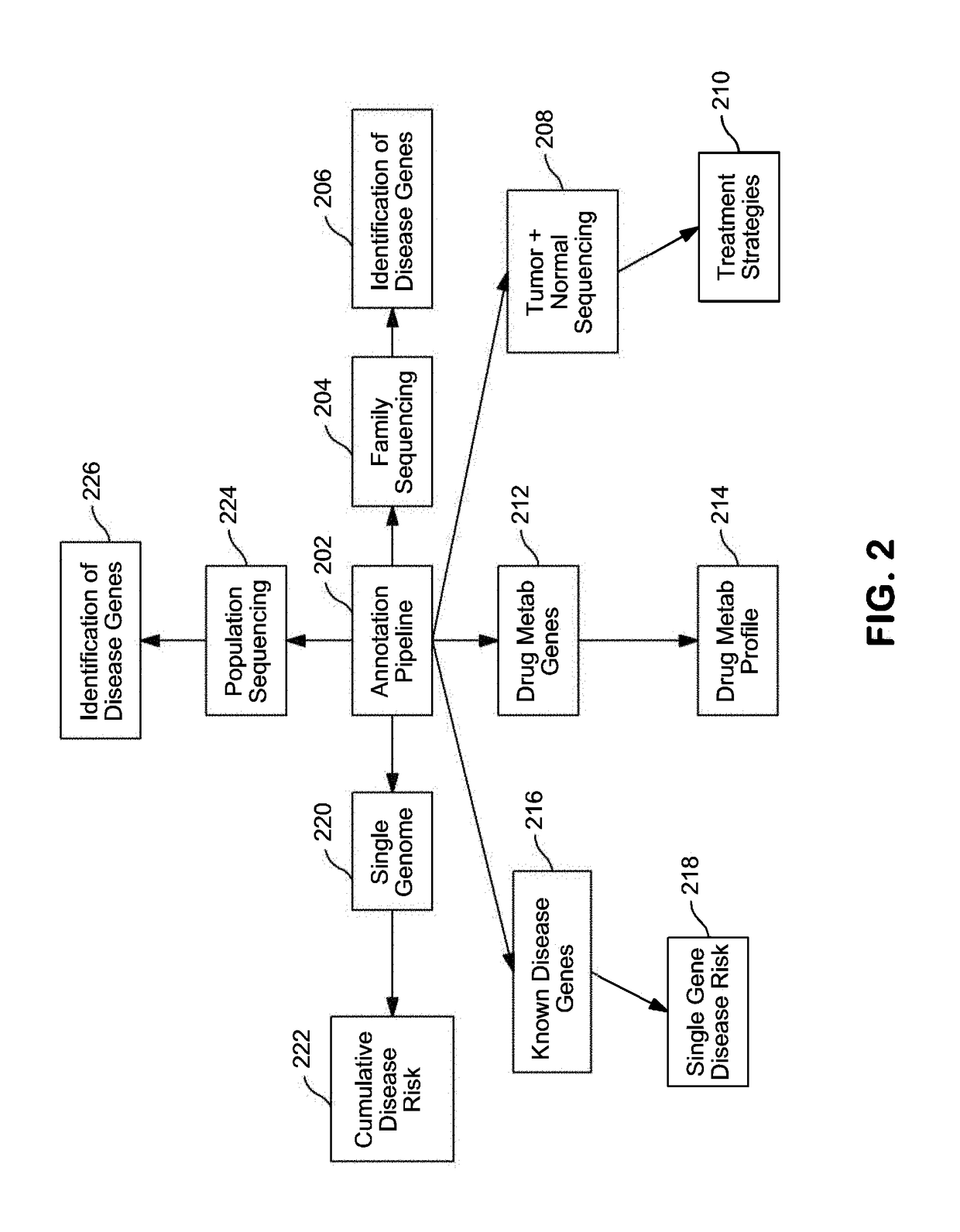
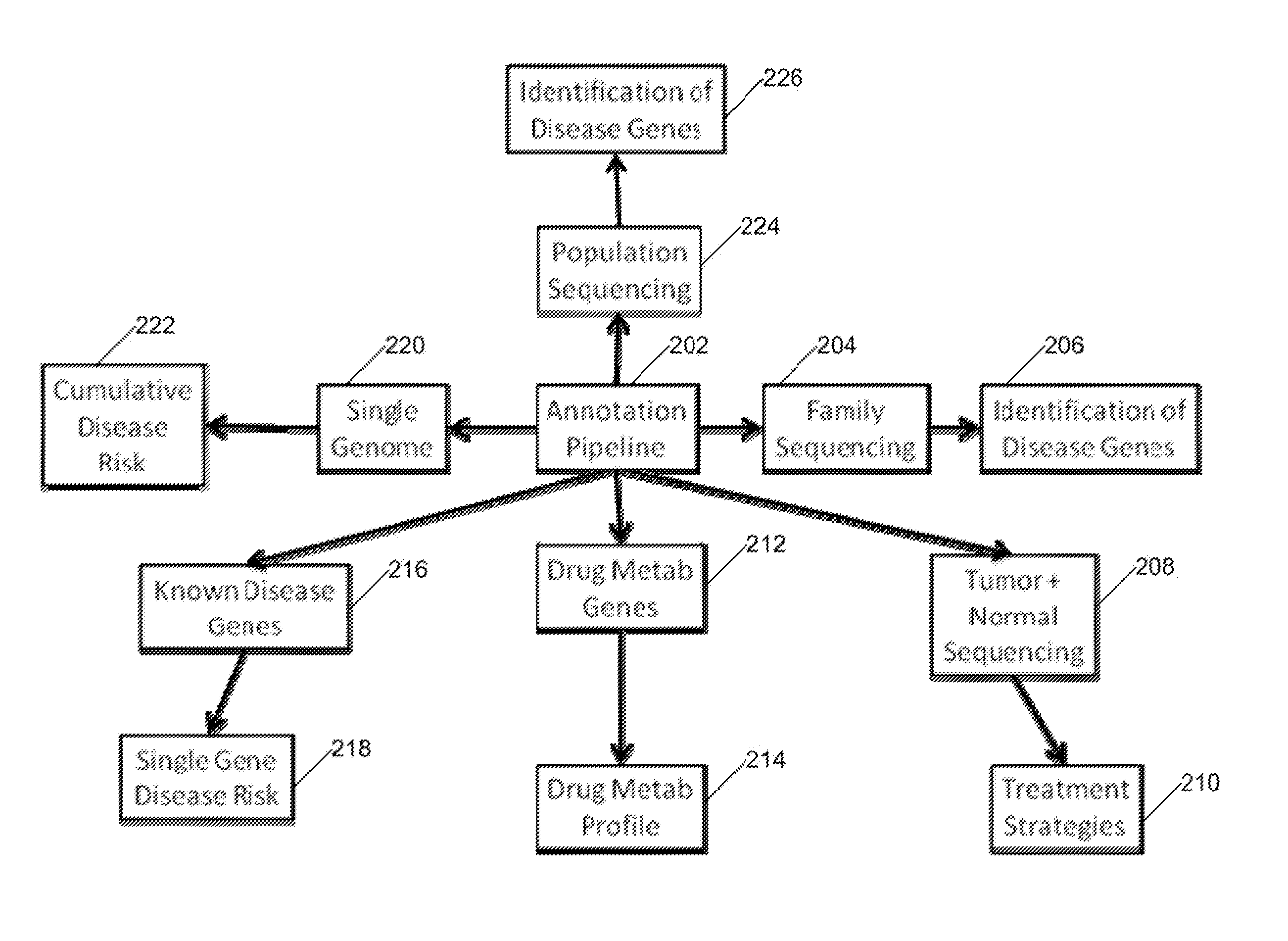
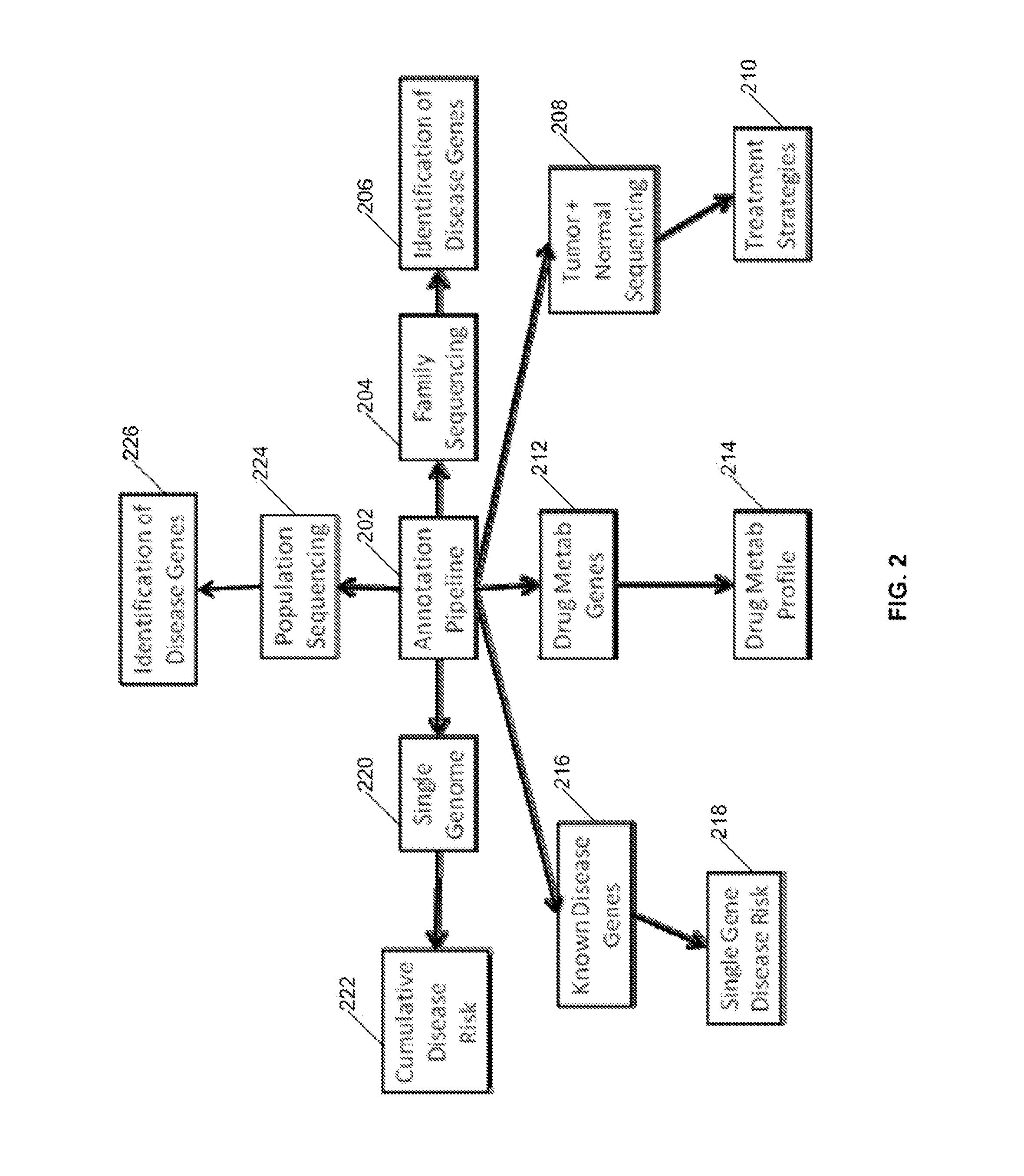
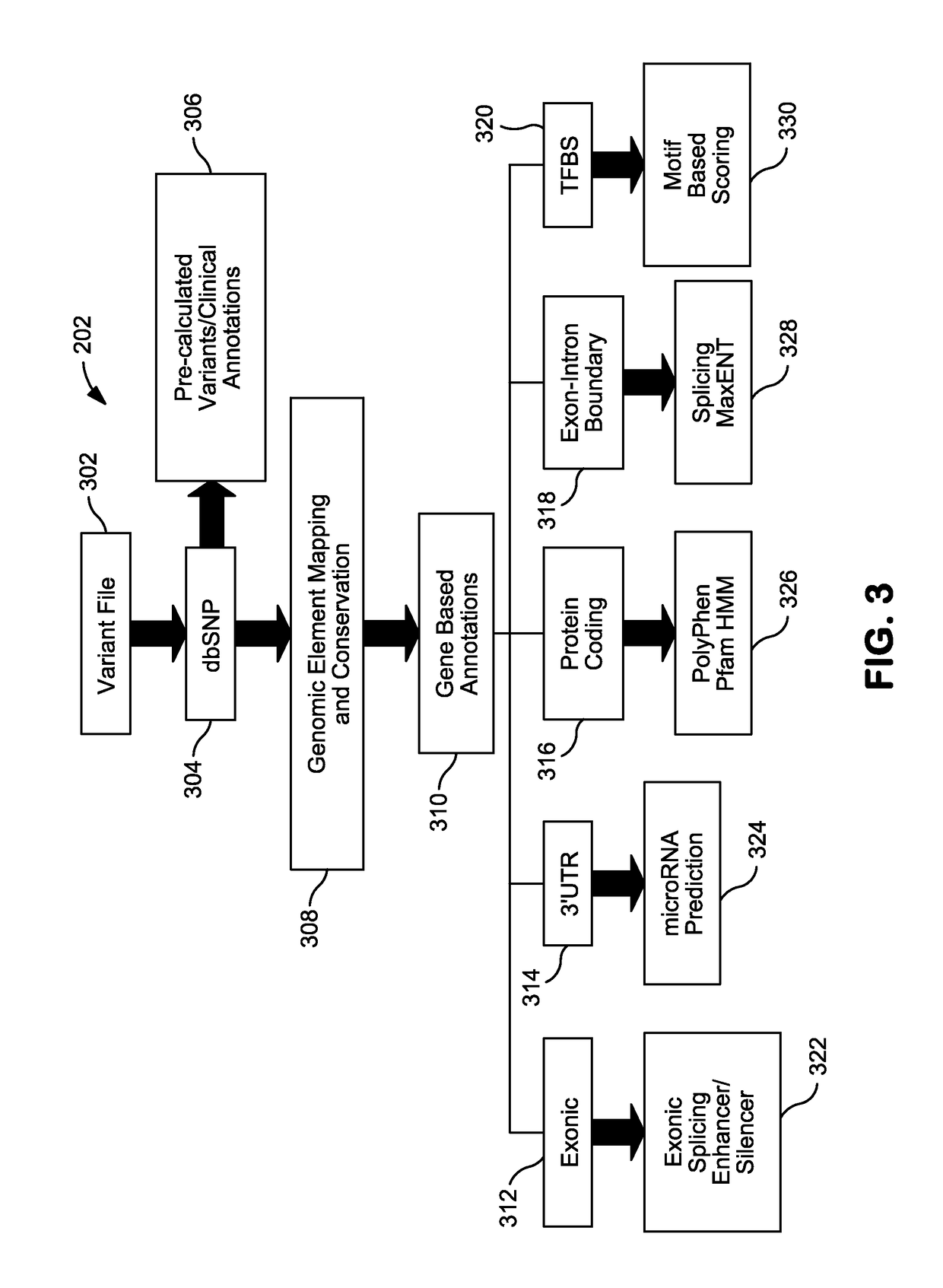
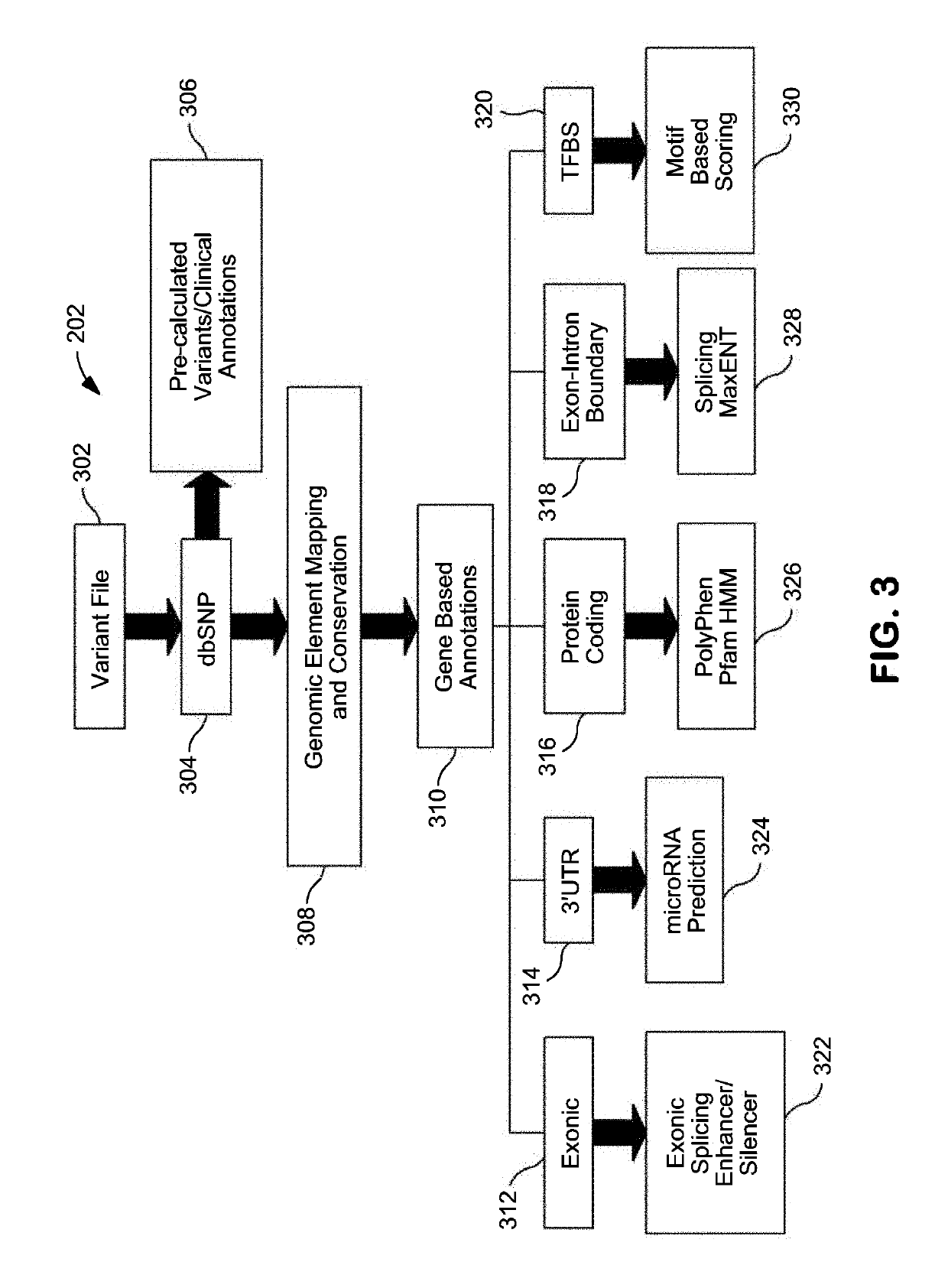
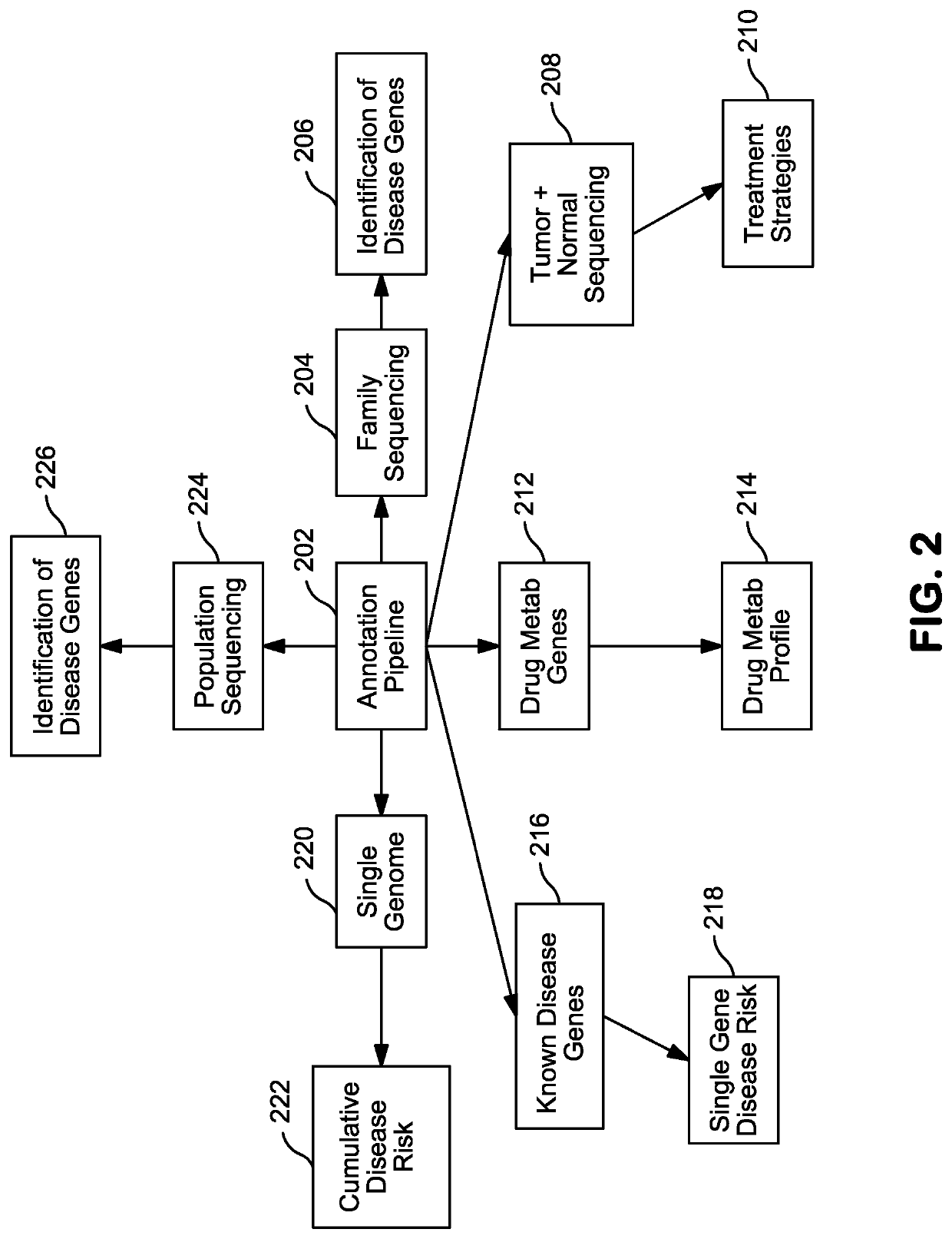
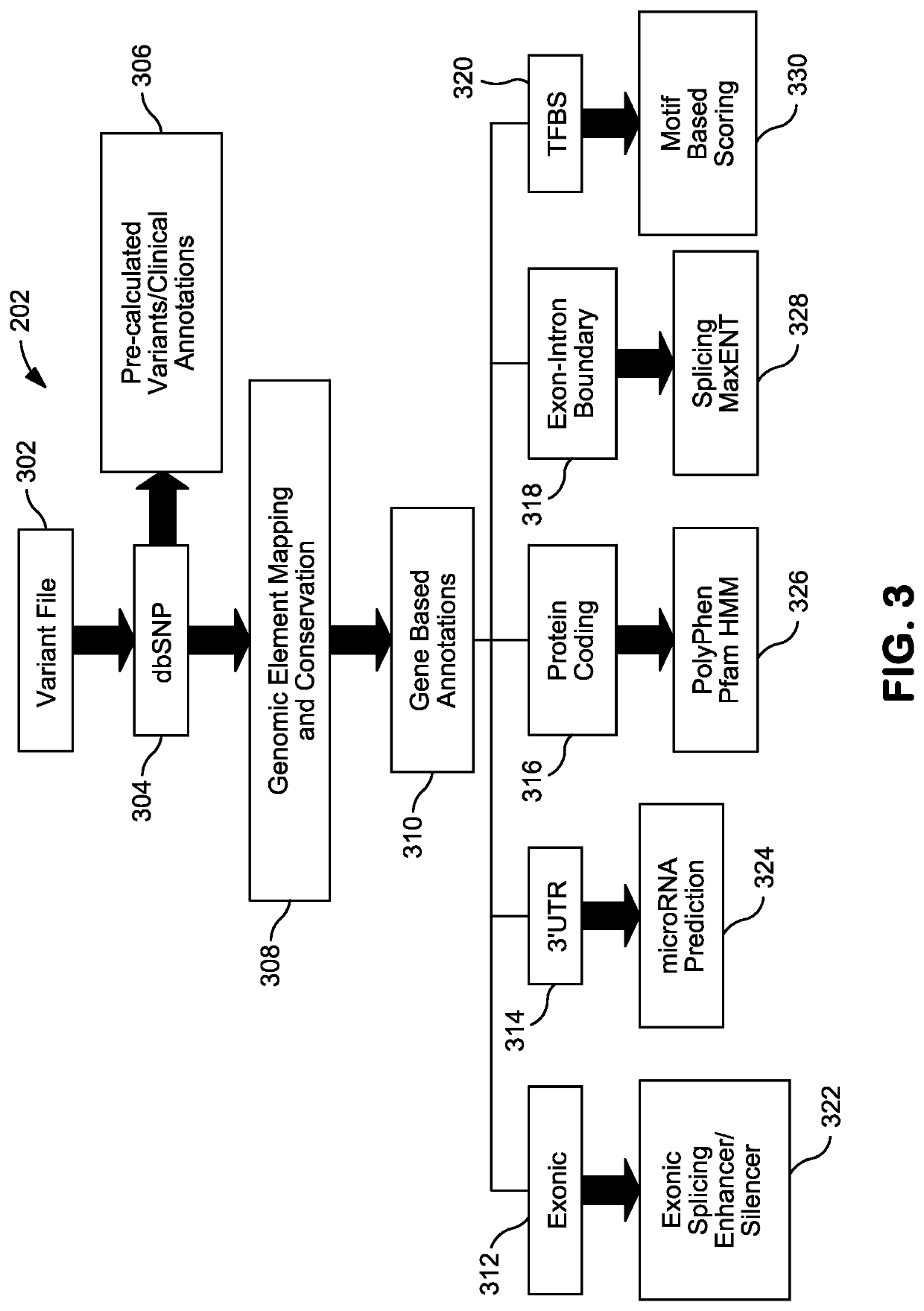
Systems and Methods for Genomic Annotation and Distributed Variant Interpretation
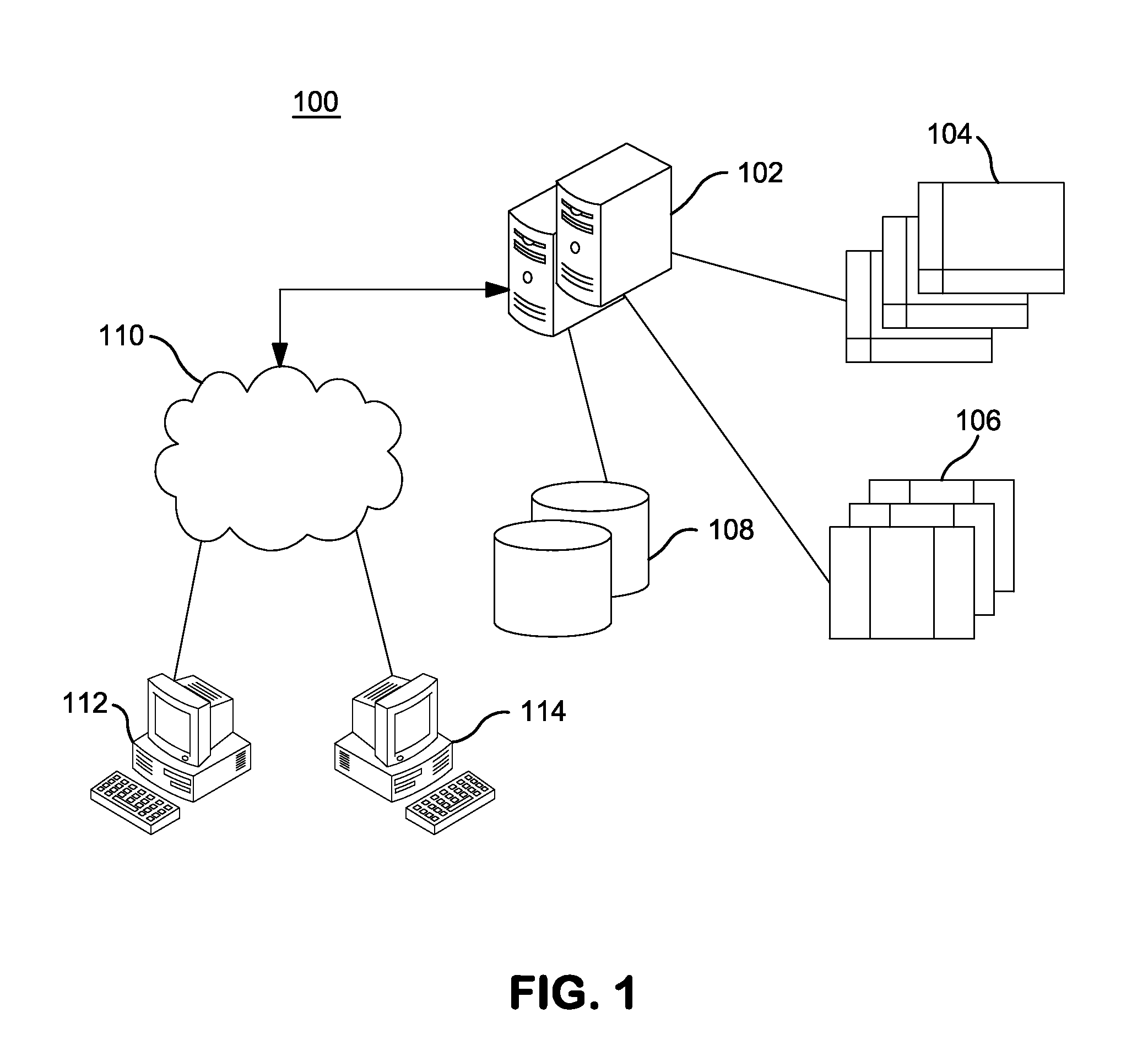
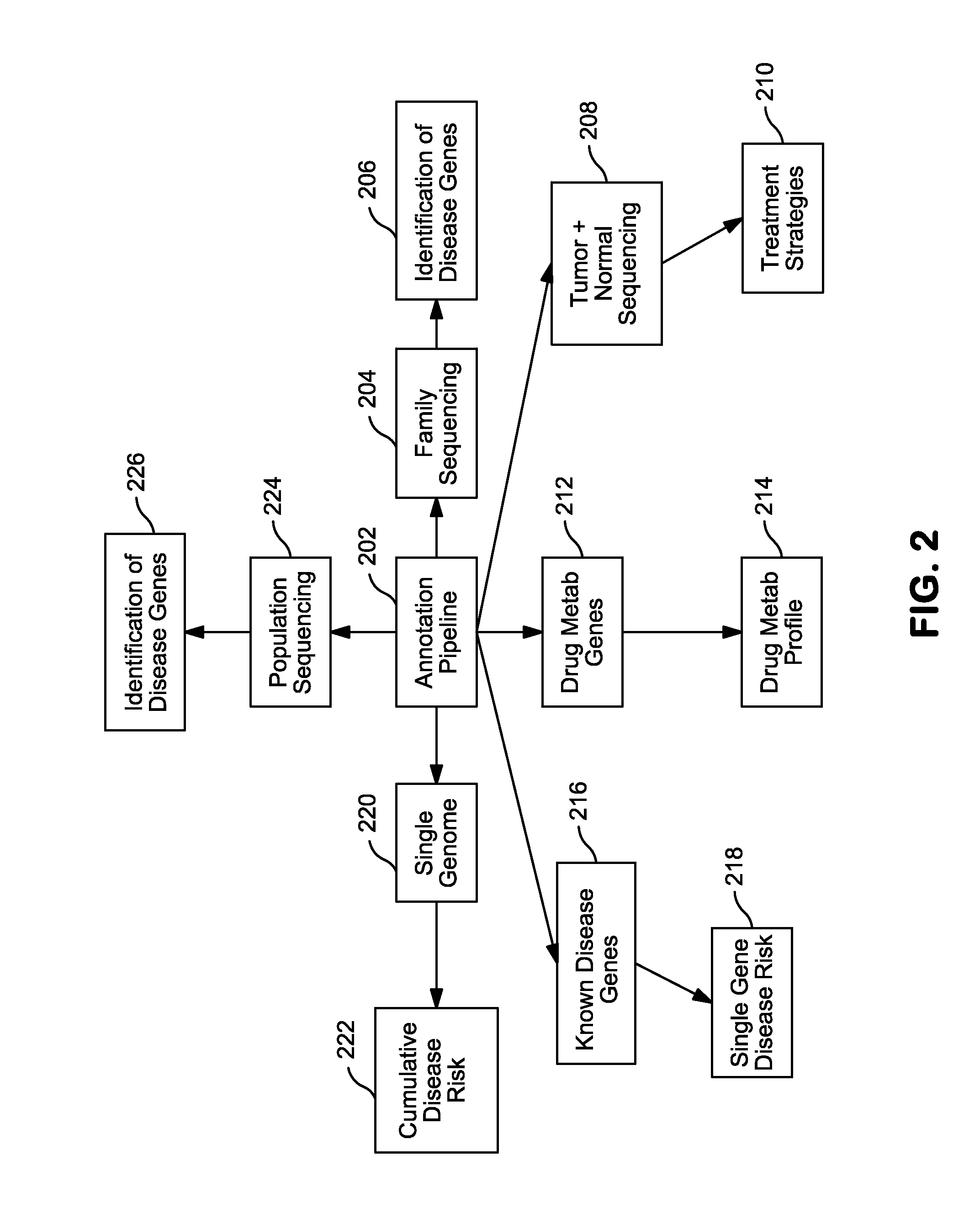
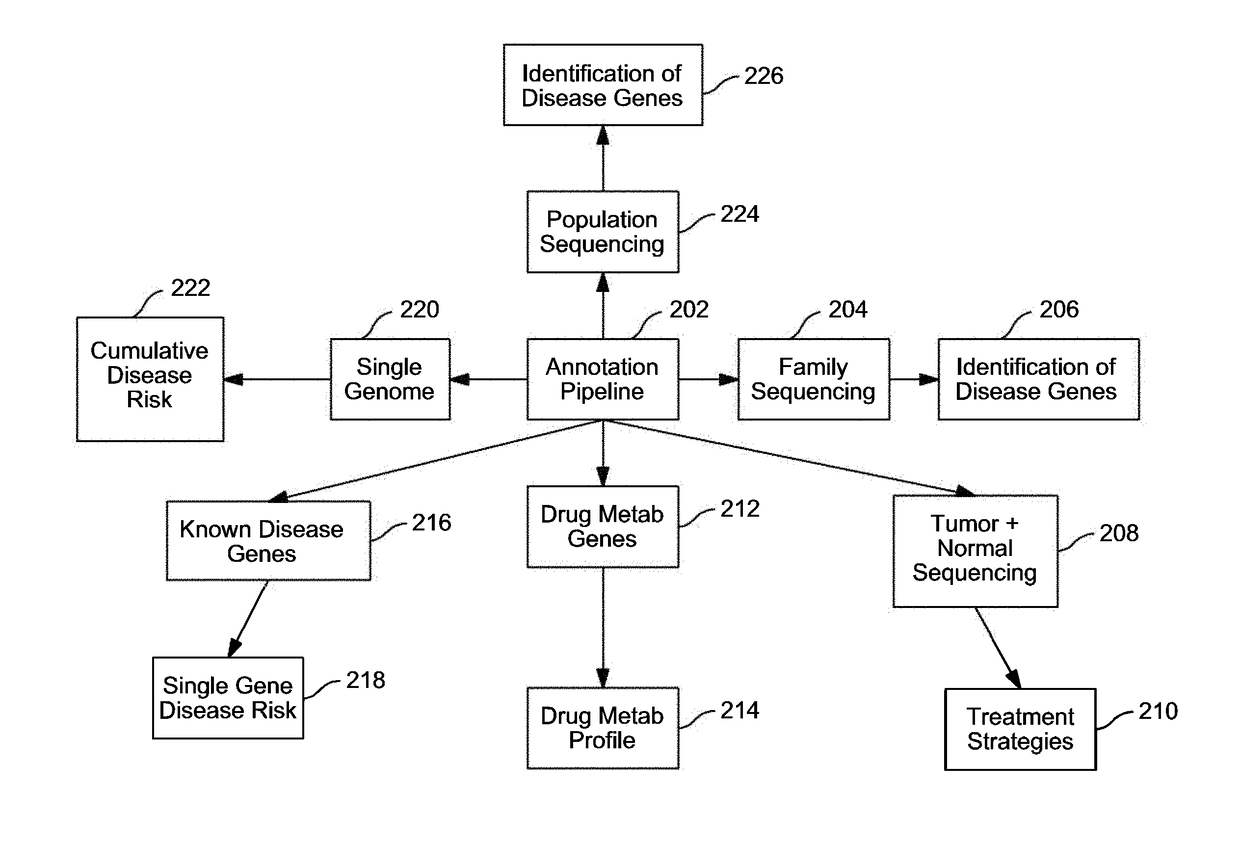
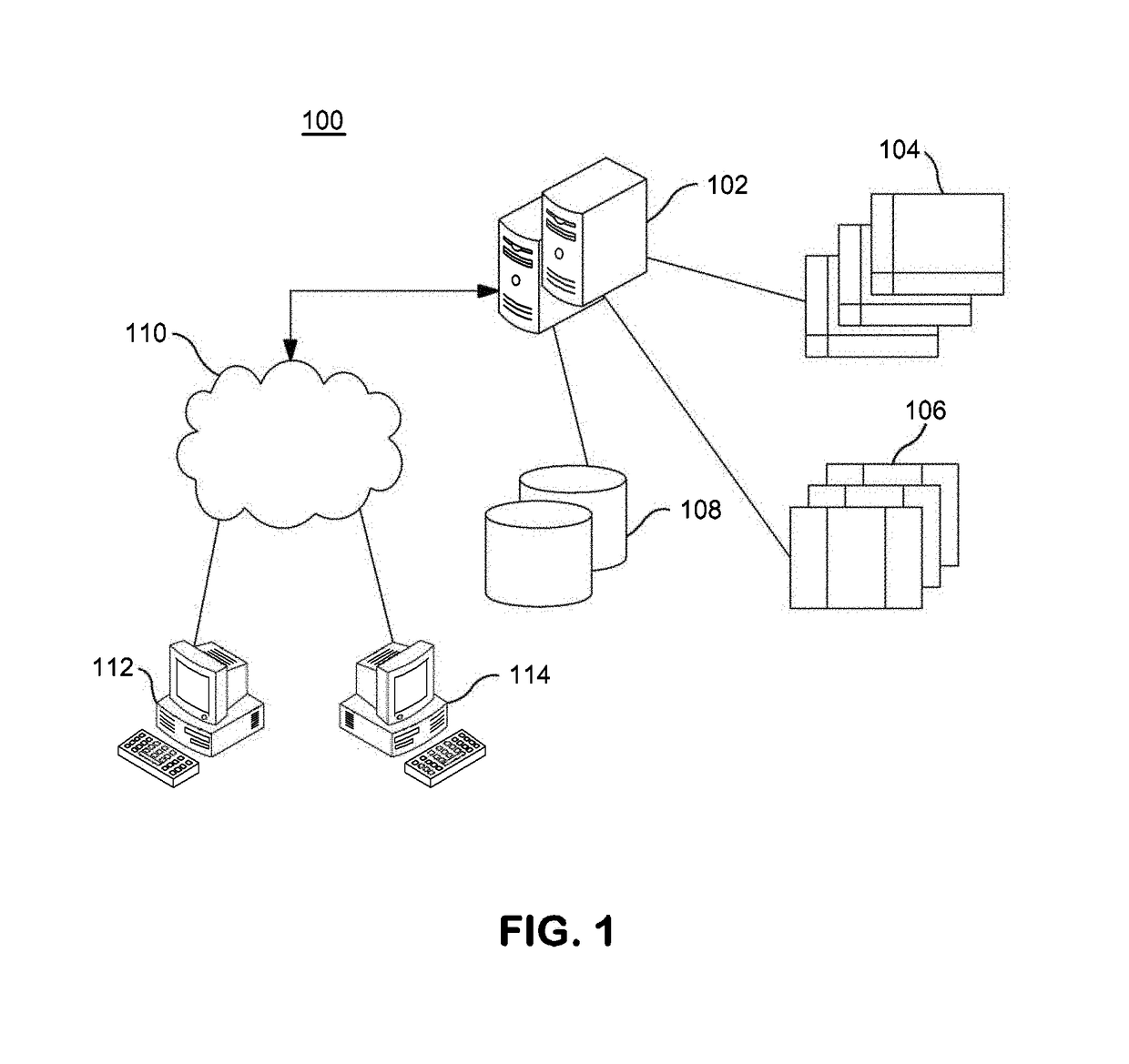
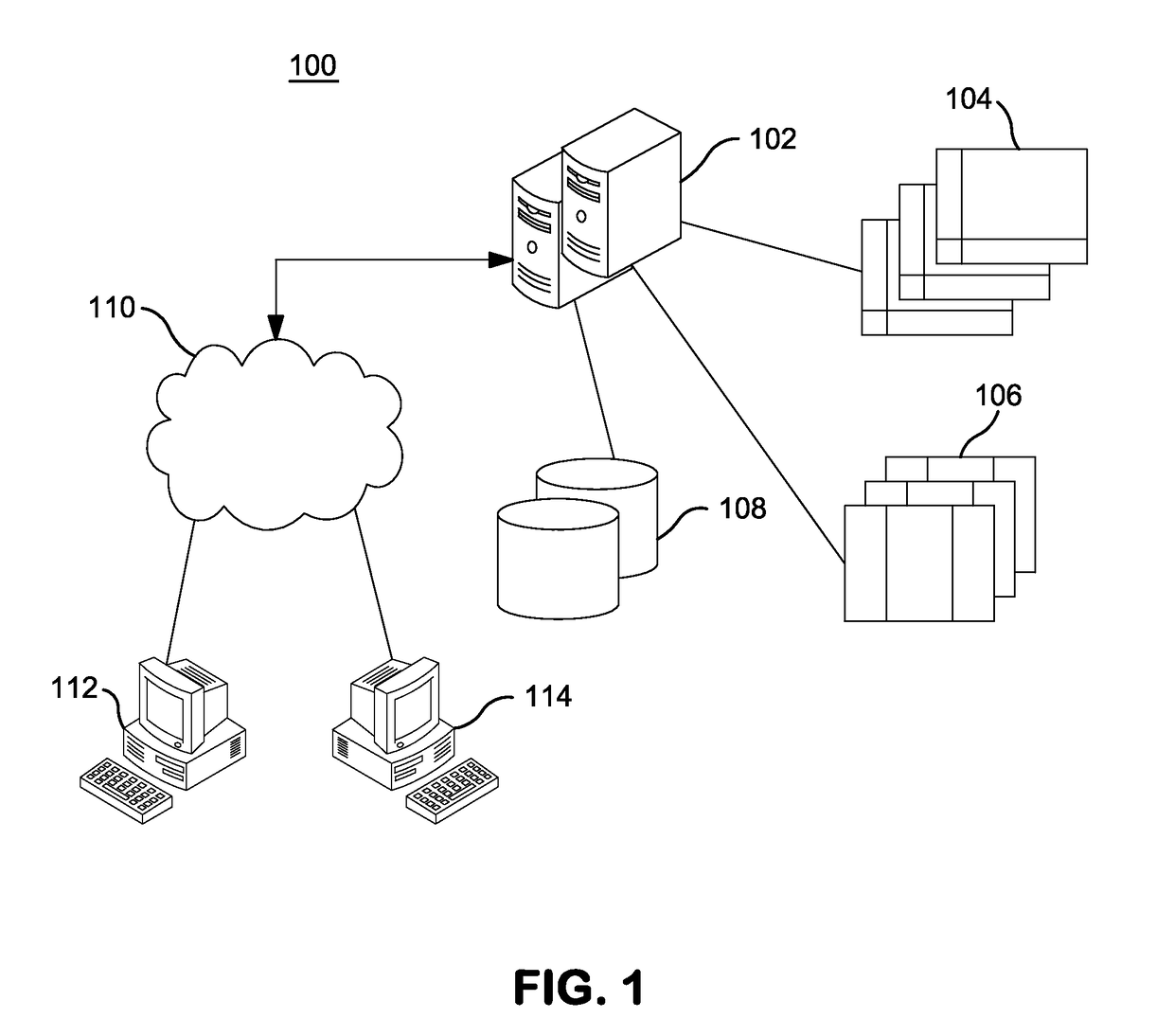
InactiveUS20140304270A1Digital data processing detailsRelational databasesGenomic dataGenomic annotation
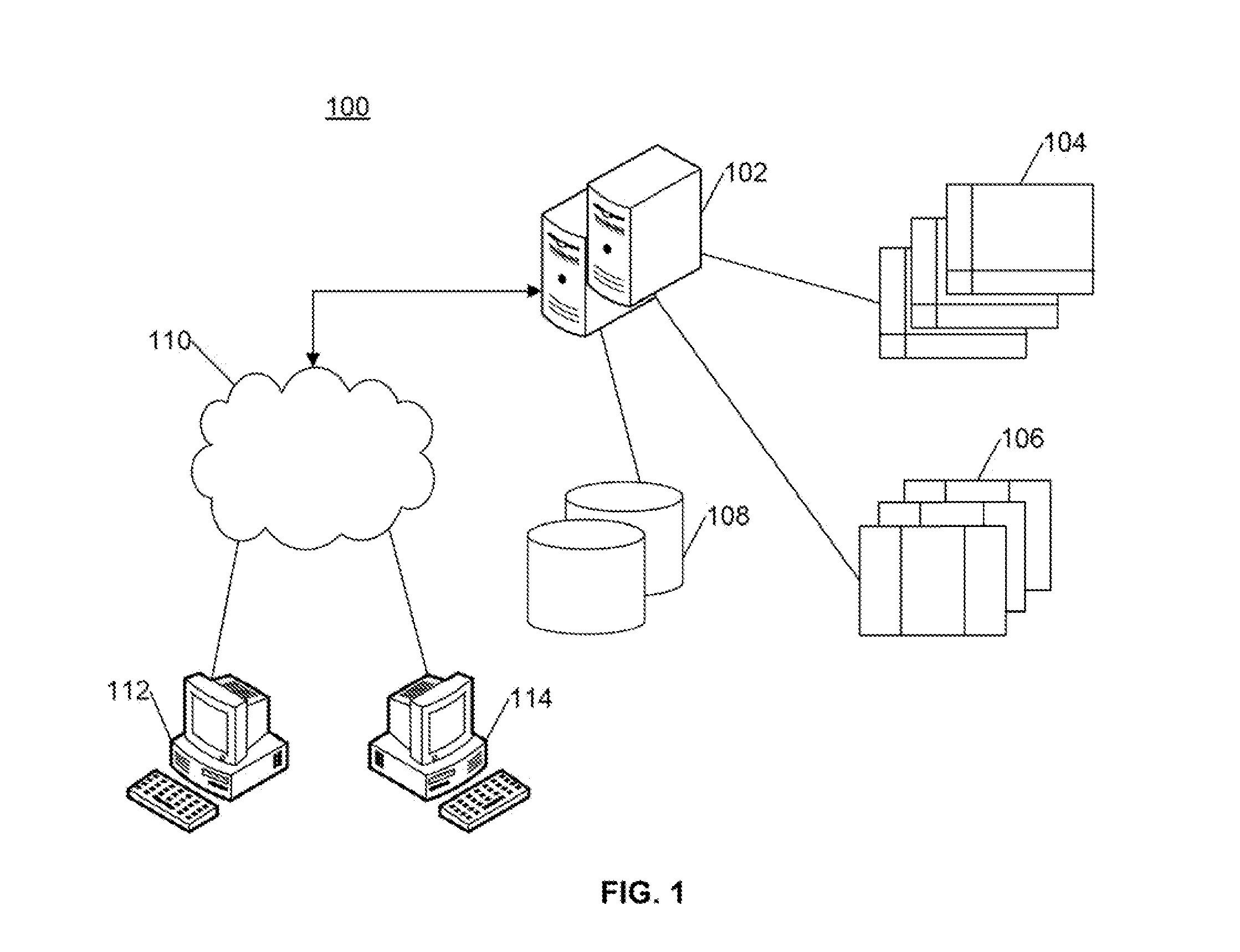
A computer-based genomic annotation system, including a database configured to store genomic data, non-transitory memory configured to store instructions, and at least one processor coupled with the memory, the processor configured to implement the instructions in order to implement an annotation pipeline and at least one module filtering or analysis of the genomic data.
Owner:THE SCRIPPS RES INST
Systems and methods for genomic annotation and distributed variant interpretation
A computer-based genomic annotation system, including a database configured to store genomic data, non-transitory memory configured to store instructions, and at least one processor coupled with the memory, the processor configured to implement the instructions in order to implement an annotation pipeline and at least one module filtering or analysis of the genomic data.
Owner:THE SCRIPPS RES INST
Systems and methods for genomic annotation and distributed variant interpretation
A computer-based genomic annotation system, including a database configured to store genomic data, non-transitory memory configured to store instructions, and at least one processor coupled with the memory, the processor configured to implement the instructions in order to implement an annotation pipeline and at least one module filtering or analysis of the genomic data.
Owner:THE SCRIPPS RES INST
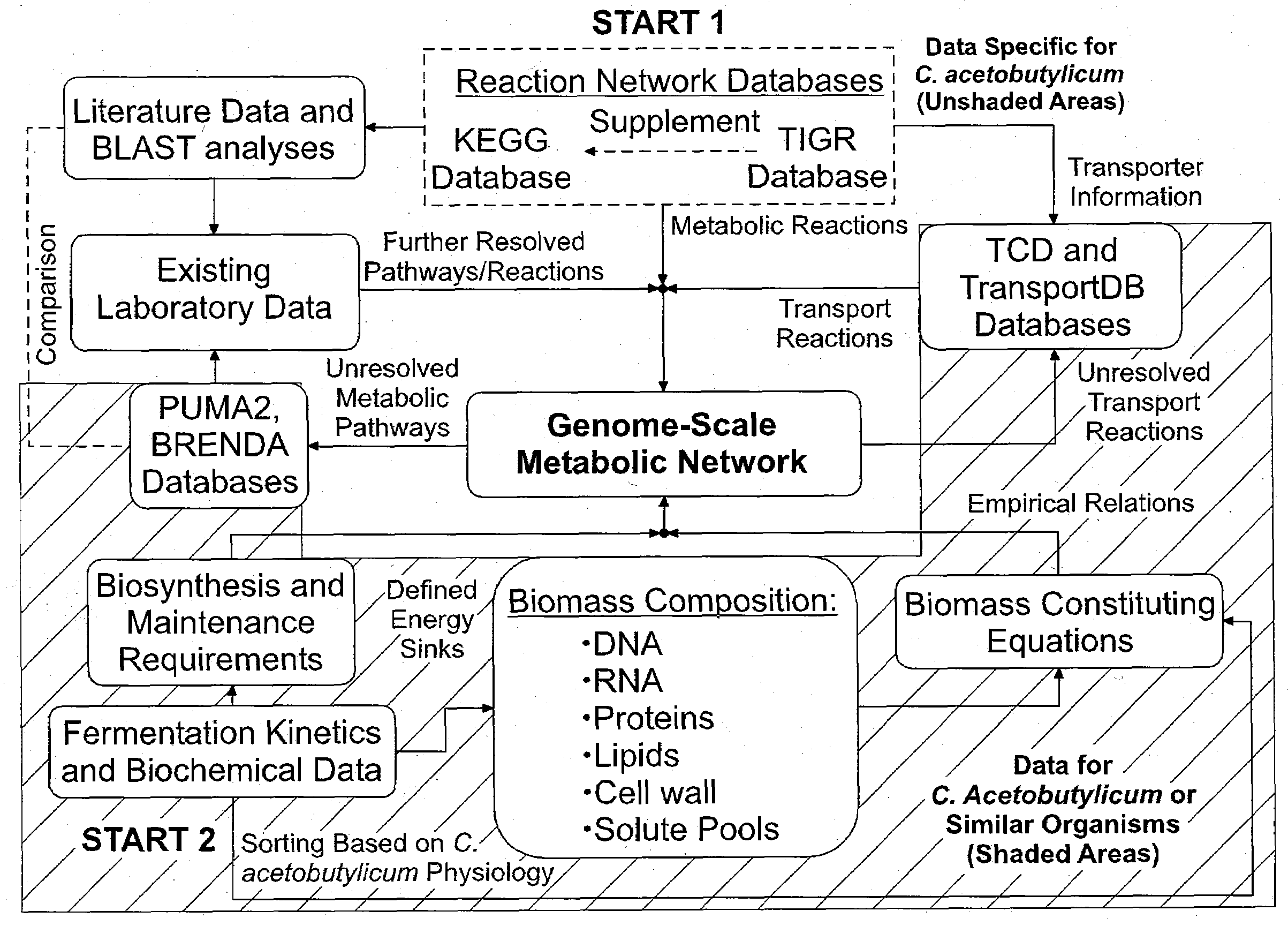
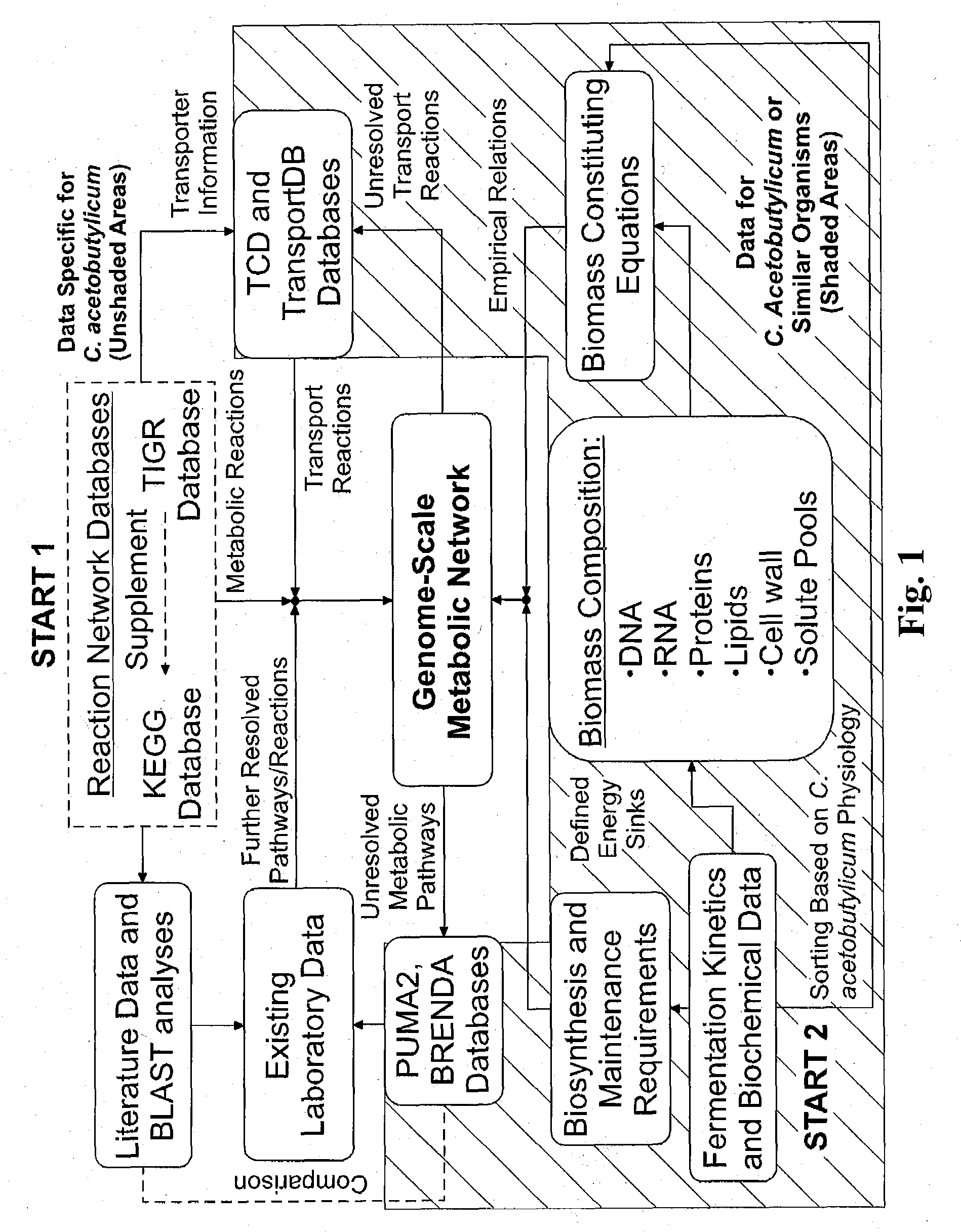
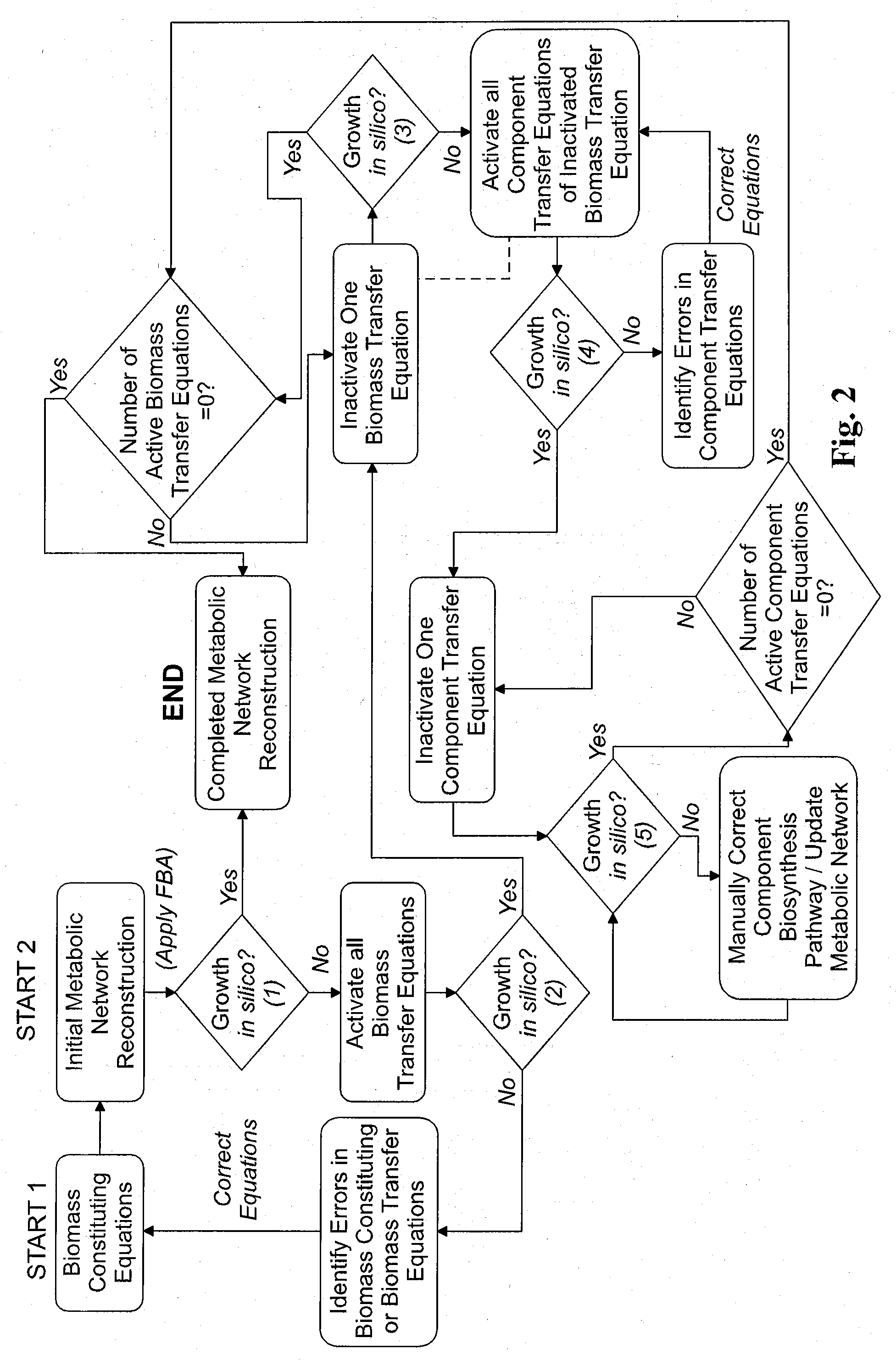
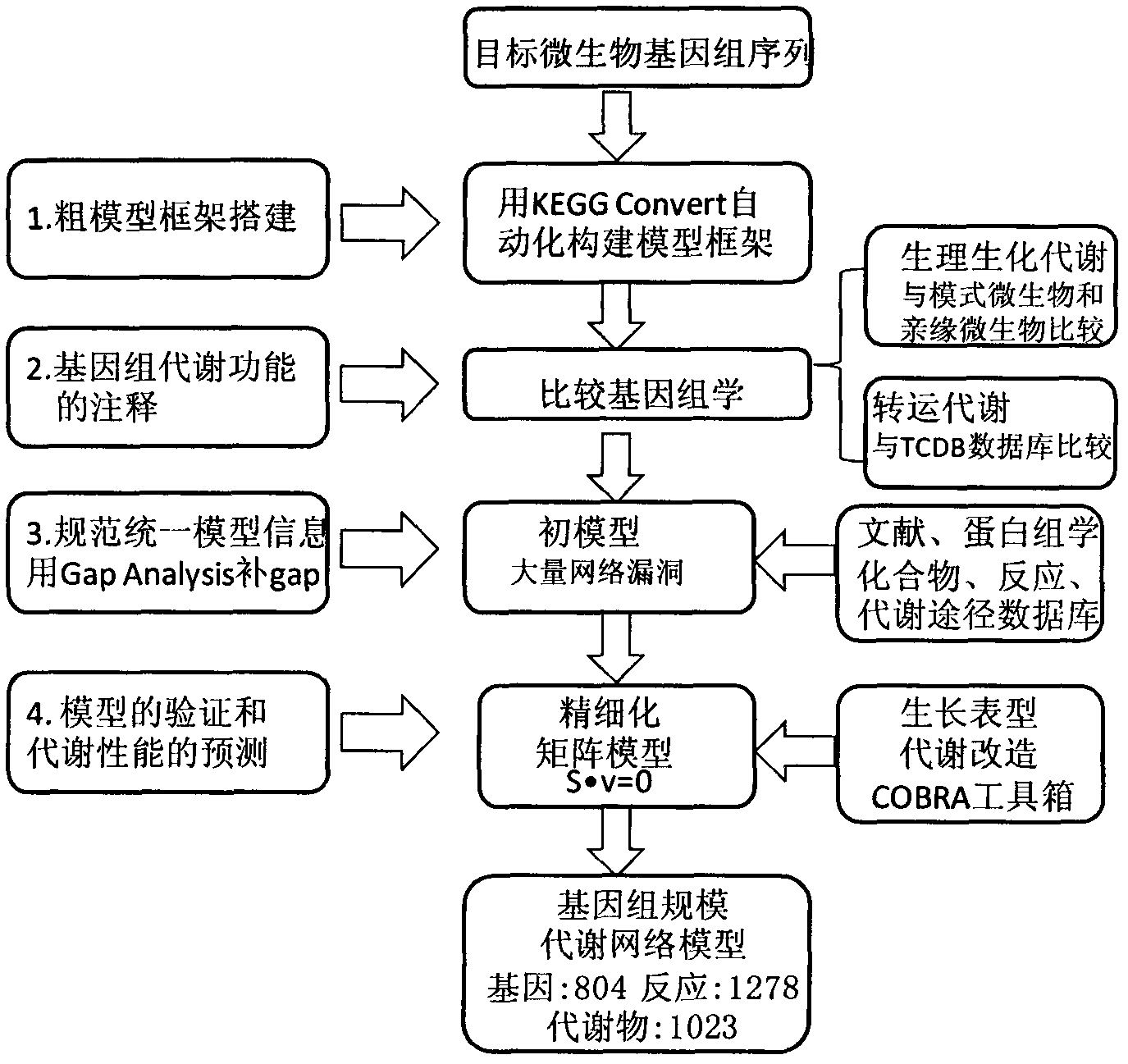
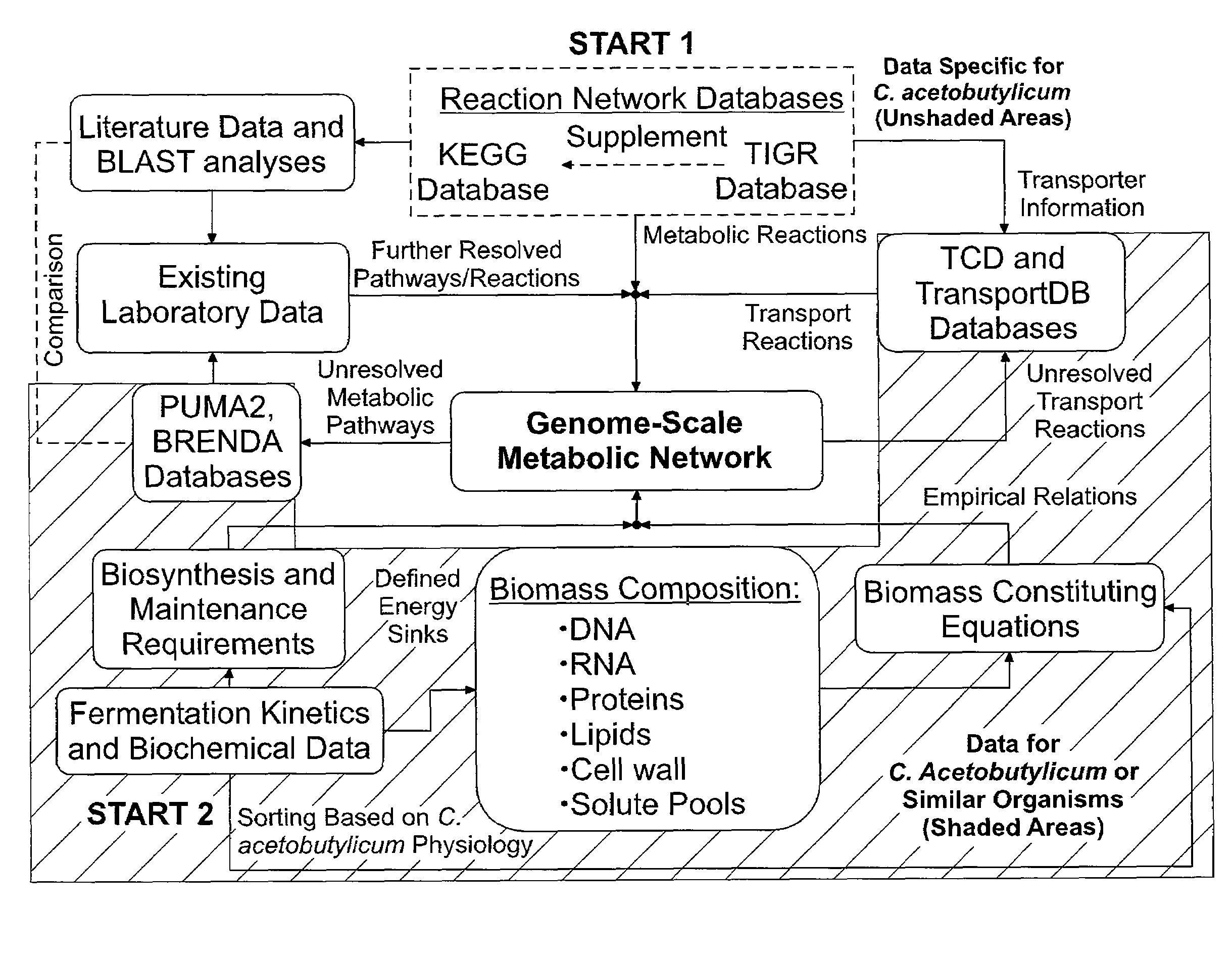
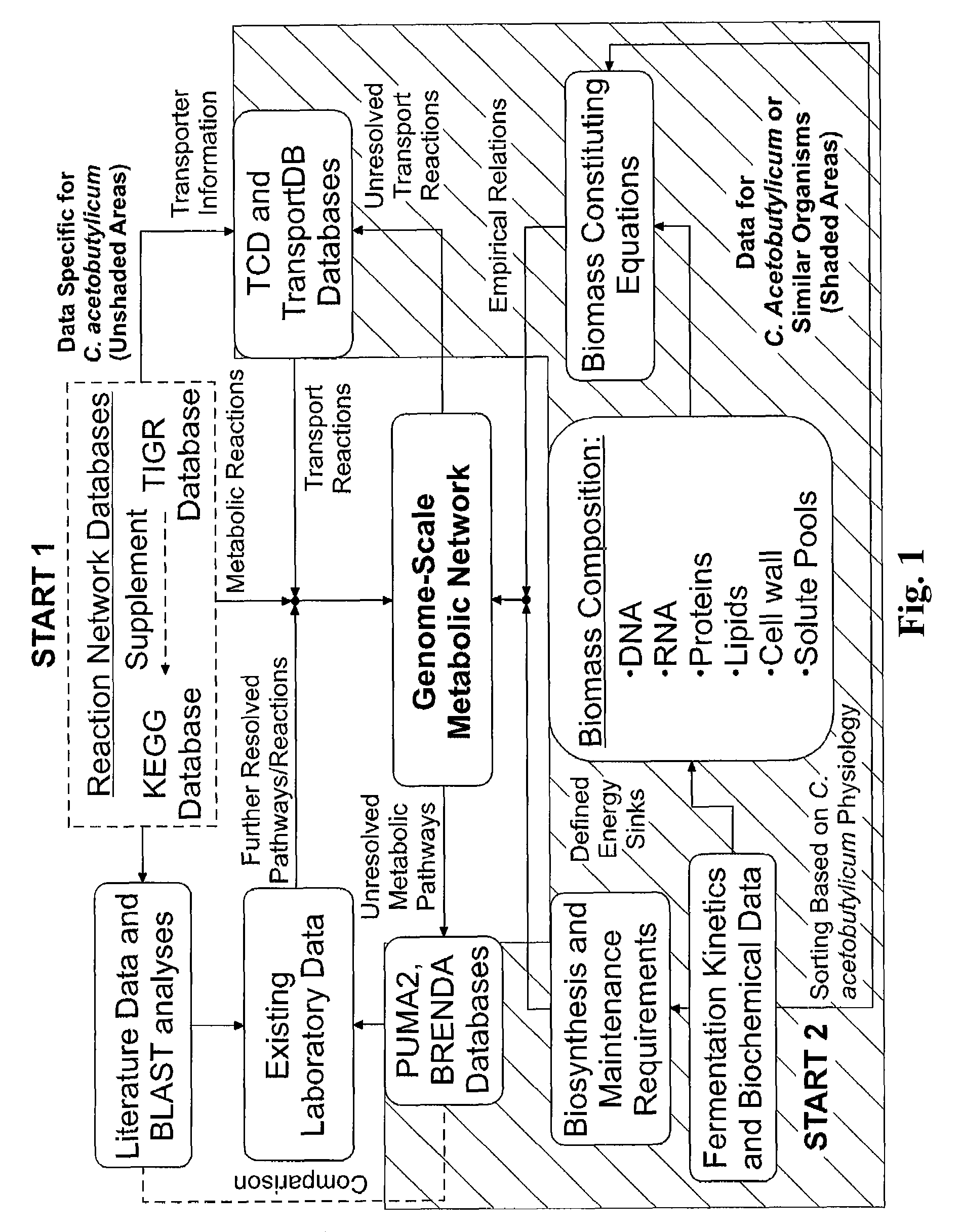
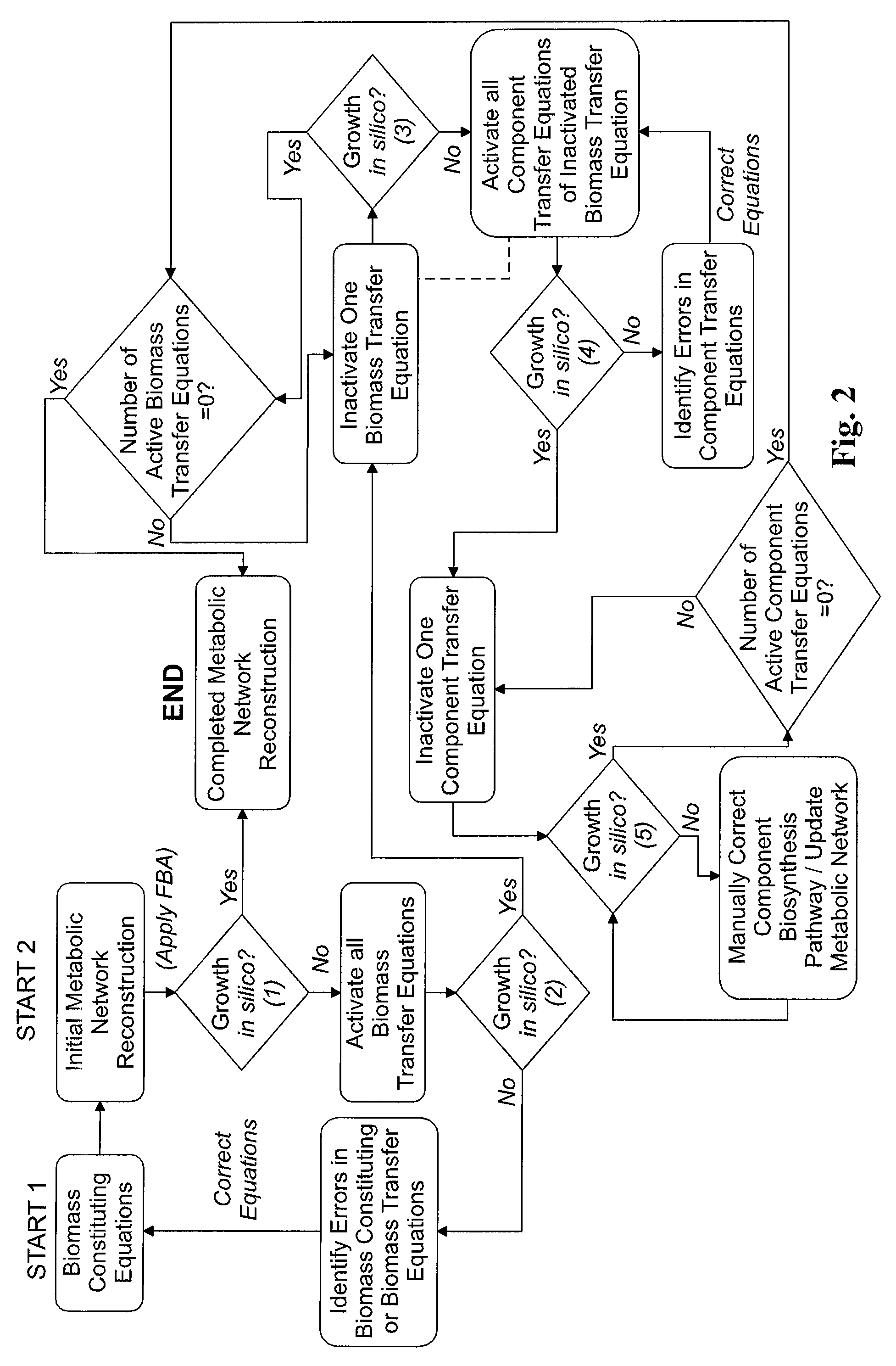
Reverse engineering genome-scale metabolic network reconstructions for organisms with incomplete genome annotation and developing constraints using proton flux states and numerically-determined sub-systems
InactiveUS20090259451A1Reduce in quantityAnalogue computers for chemical processesBiological testingUnderdetermined systemIn silico
A genome-scale metabolic network reconstruction for Clostridium acetobutylicum (ATCC 824) was created using a new semi-automated reverse engineering algorithm. This invention includes algorithms and software that can reconstruct genome-scale metabolic networks for cell-types available through the Kyoto Encyclopedia of Genes and Genomes. This method can also be used to complete partial metabolic networks and cell signaling networks where adequate starting information base is available. The software may use a semi-automated approach which uses a priori knowledge of the cell-type from the user. Upon completion, the program output is a genome-scale stoichiometric matrix capable of cell growth in silico. The invention also includes methods for developing flux constraints and reducing the number of possible solutions to an under-determined system by applying specific proton flux states and identifying numerically-determined sub-systems. Although the model-building and analysis tools described in this invention were initially applied to C. acetobutylicum, the novel algorithms and software can be applied universally.
Owner:UNIVERSITY OF DELAWARE
Systems and Methods for Genomic Annotation and Distributed Variant Interpretation
A computer-based genomic annotation system, including a database configured to store genomic data, non-transitory memory configured to store instructions, and at least one processor coupled with the memory, the processor configured to implement the instructions in order to implement an annotation pipeline and at least one module filtering or analysis of the genomic data.
Owner:THE SCRIPPS RES INST
Method for accurately predicting BSA-seq candidate gene in low-density SNP genomic region
ActiveCN109360606AMake up for false positivesMicrobiological testing/measurementProteomicsConfidence intervalCandidate Gene Association Study
The invention relates to the genome sequencing technology field and especially relates to a method for accurately predicting a BSA-seq candidate gene in a low-density SNP genomic region. BSA-seq has alow-density SNP region near a candidate interval. Through comparing SNPs between two parents, an SNP list is strictly filtered to find a low-density region, and then, a corresponding candidate interval when a confidence interval is 95% and a low-density candidate interval are used, and a genome annotation website is used to annotate the genes in a candidate region. A candidate region variation point function is annotated so as to obtain a gene with functional variation such as frameshift variation and the like, and the gene is determined to be a candidate gene. The method can be used to makeup for the false positive of the candidate region due to the region with a small difference in a genome, and the real candidate interval is acquired.
Owner:广西壮族自治区农业科学院水稻研究所
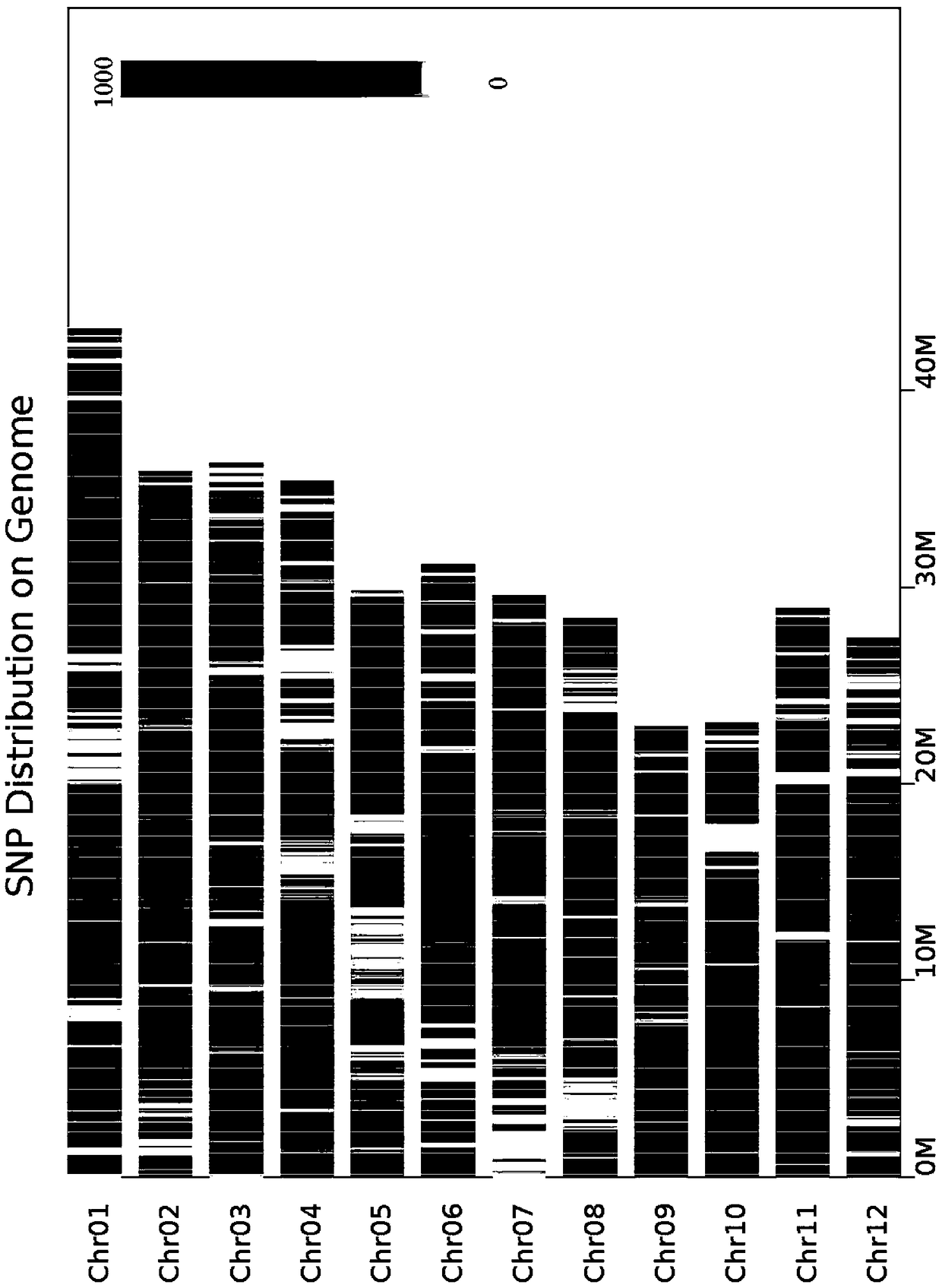
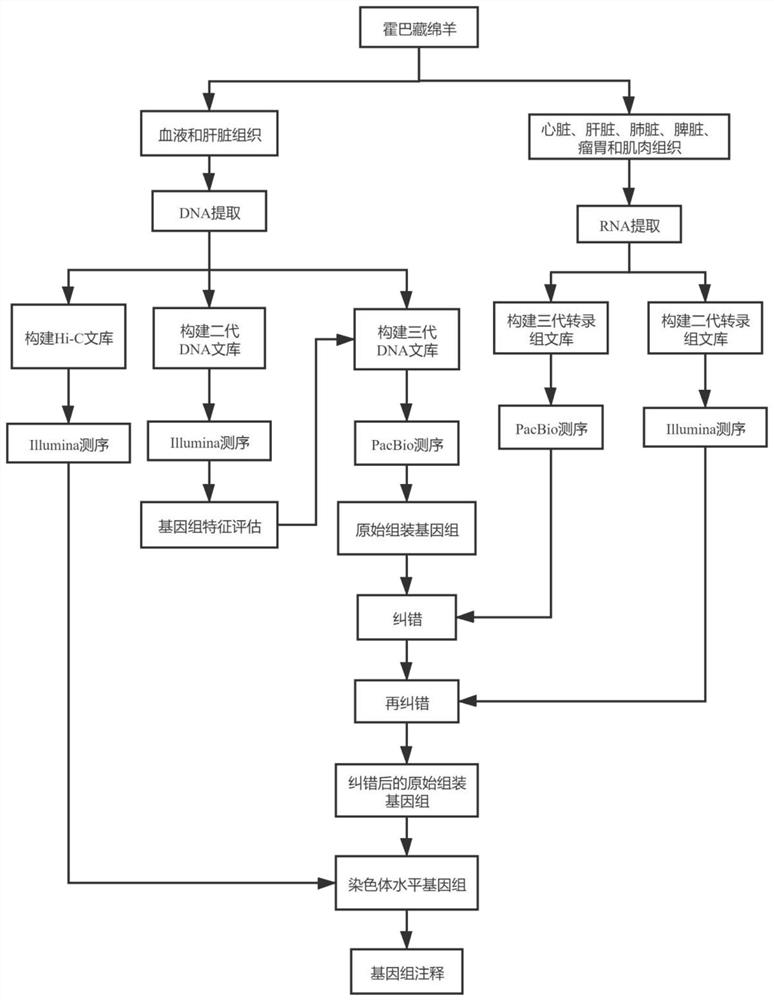
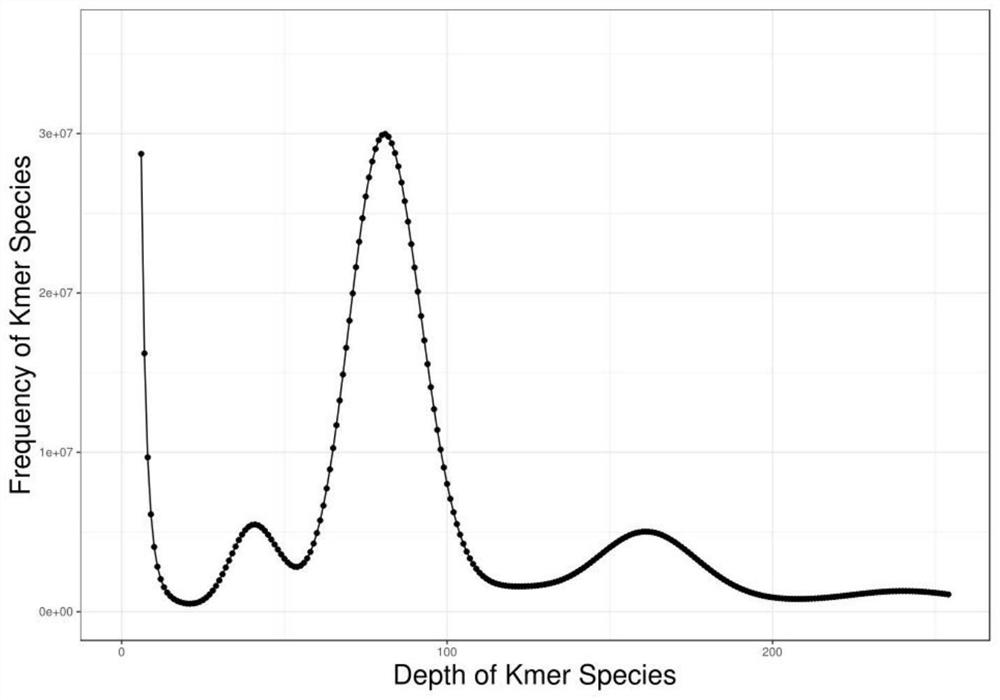
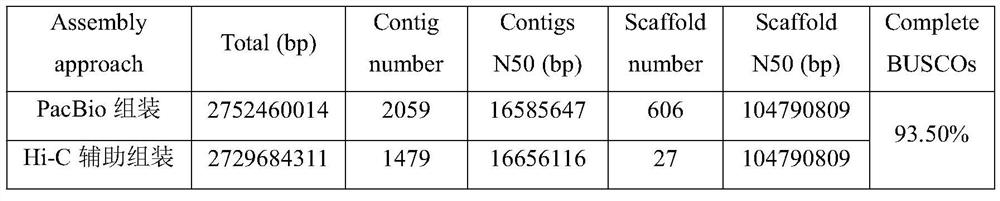
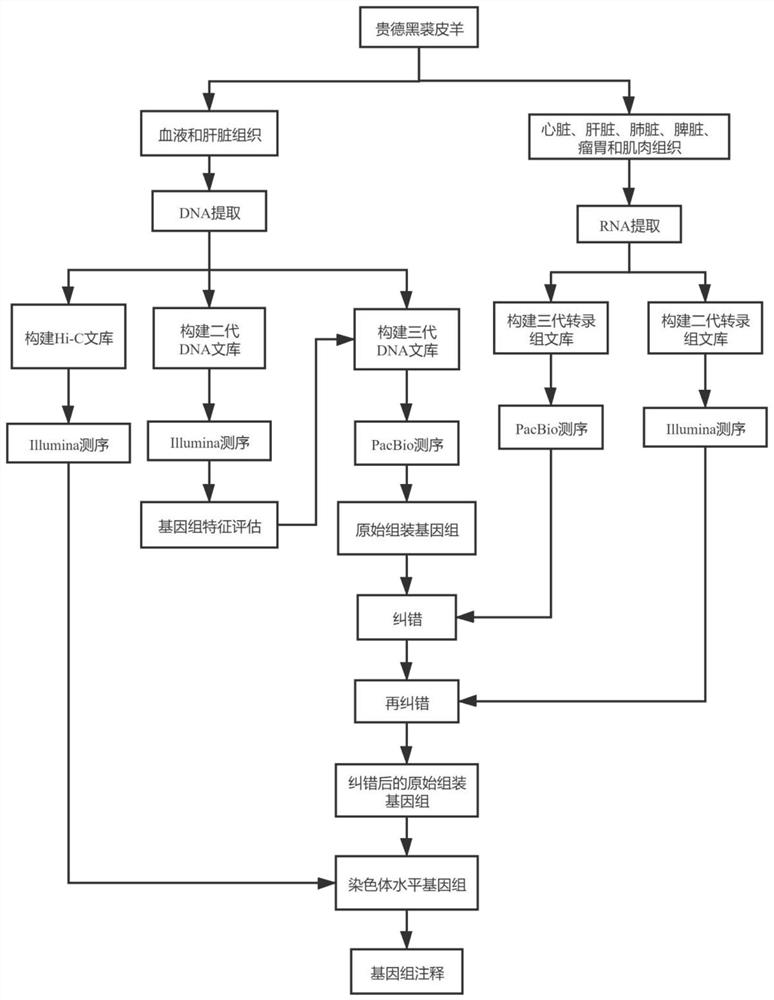
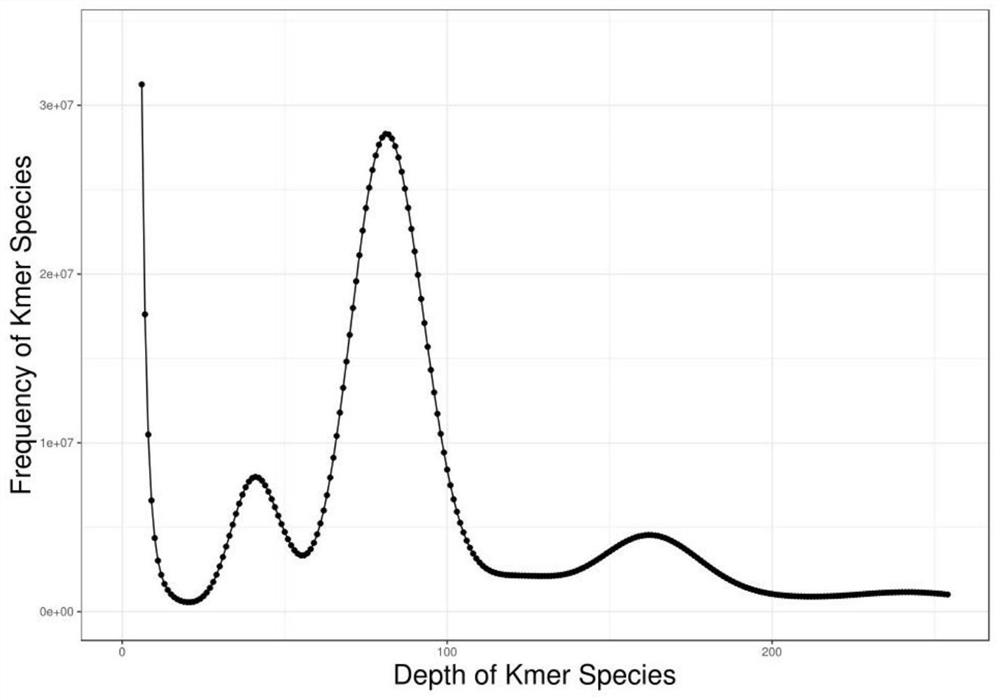
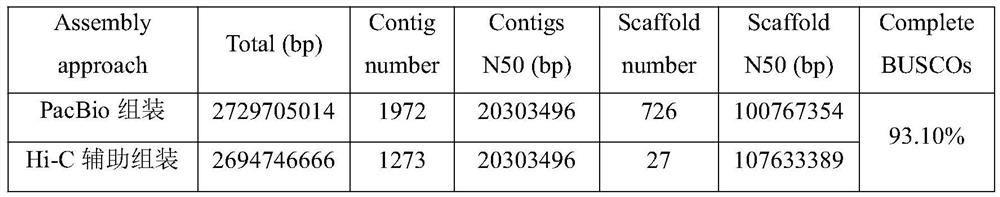
Method for assembling and annotating genome of Hoba Tibetan sheep based on three-generation PacBio and Hi-C technologies
PendingCN113151426AImprove integrityImprove continuityMicrobiological testing/measurementProteomicsBiotechnologyGenetics
The invention belongs to the technical field of bioinformatics, and particularly relates to a method for assembling and annotating a genome of Hoba Tibetan sheep based on three-generation PacBio and Hi-C technologies. The method comprises the following steps: (1) collecting blood and tissue samples of Hoba Tibetan sheep; (2) constructing a genome library and a transcriptome library; (3) evaluating genome size and heterozygosity rate; (4) assembling genome, conducting error correction by utilizing a transcription sequencing result; (5) conducting Hi-C assisted assembly and evaluation; and (6) conducting genome annotation and evaluation. According to the method, the high-quality genome of the Hoba Tibetan sheep is assembled at a chromosome level, thereby providing valuable genome resources for studying conservation and innovative utilization of genetic resources of Tibetan sheep populations, and laying a solid foundation for further researches on an environment adaptation mechanism of specific livestock species in the Qinghai-Tibet Plateau. The method of the present invention lays foundation for researches on an alpine and hypoxia adaptive molecular mechanism of the Hoba Tibetan sheep, and provides reference data for researches on human hypoxia-related diseases.
Owner:LANZHOU INST OF ANIMAL SCI & VETERINARY PHARMA OF CAAS
Construction system of novel transposon mutant strain library
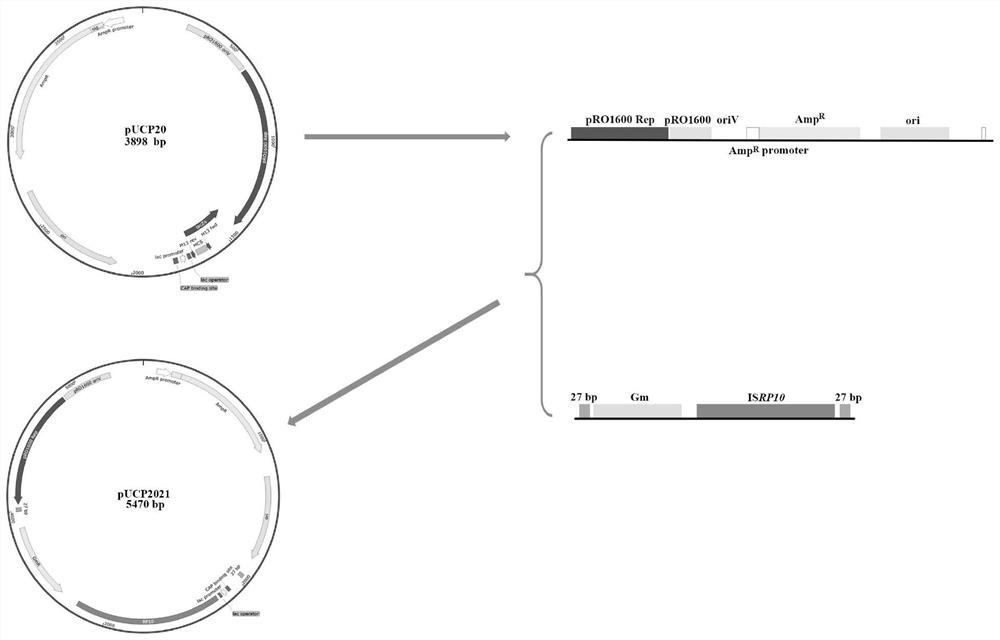
ActiveCN112626064AIncrease diversityReduce capacityBacteriaMicroorganism based processesPlasmid VectorInsertion sequence
Owner:TIANJIN UNIVERSITY OF SCIENCE AND TECHNOLOGY
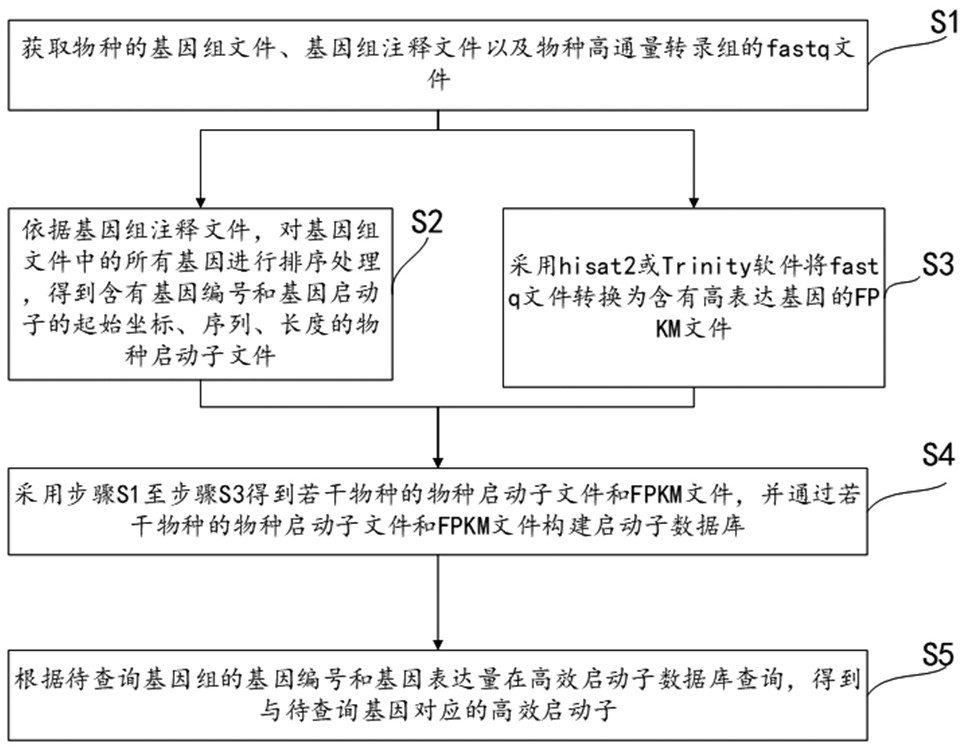
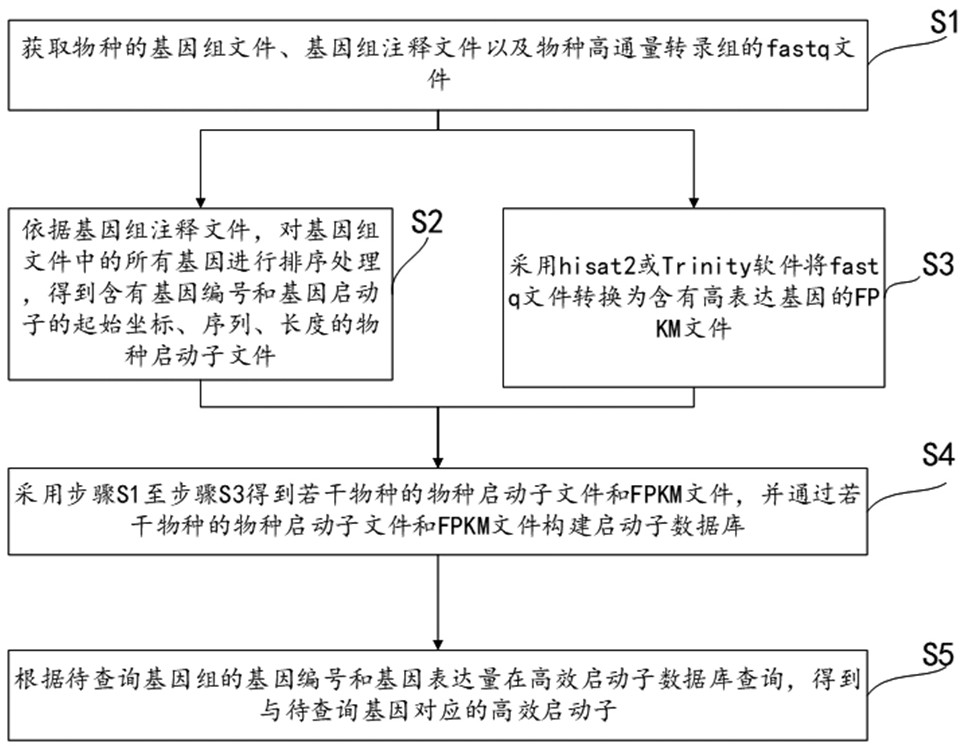
Promoter query method, system and equipment for species genome
ActiveCN113284559AImprove accuracyAdapt to the needs of transformation carriersDigital data information retrievalSequence analysisExpression geneGenomic annotation
The invention relates to a promoter query method, system and equipment for species genome. The method comprises the following steps: acquiring a genome file, a genome annotation file and a fastq file of a species; sequencing all genes in the genome file according to the genome annotation file to obtain a species promoter file; converting the fastq file into an FPKM file containing a high expression gene by adopting hisat2 or Trinity software; obtaining the species promoter files and the FPKM files of the multiple species through the steps S1 to S3, and constructing the promoter database through the species promoter files and the FPKM files of the multiple species. According to the promoter query method of the species genome, the promoter of the required gene can be queried in the promoter database, the query of the gene promoter of any species is not limited, an additional auxiliary tool is not needed, and the accuracy of the queried promoter is high.
Owner:JINAN UNIVERSITY
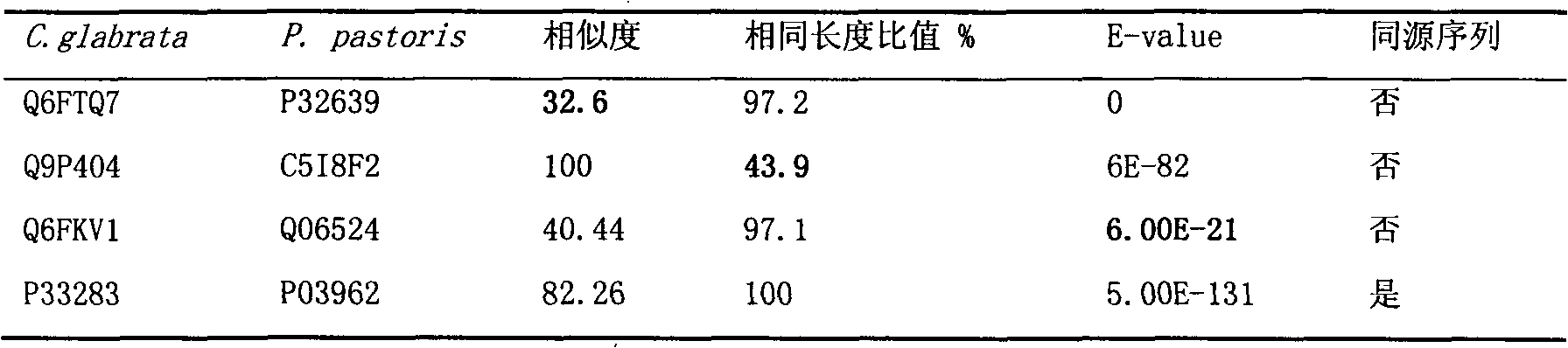
Construction and application technology of Torulopsis glabrata genome metabolism model
InactiveCN102622533AIncrease productionSpecial data processing applicationsTorulopsis glabrataEthanol dehydrogenase
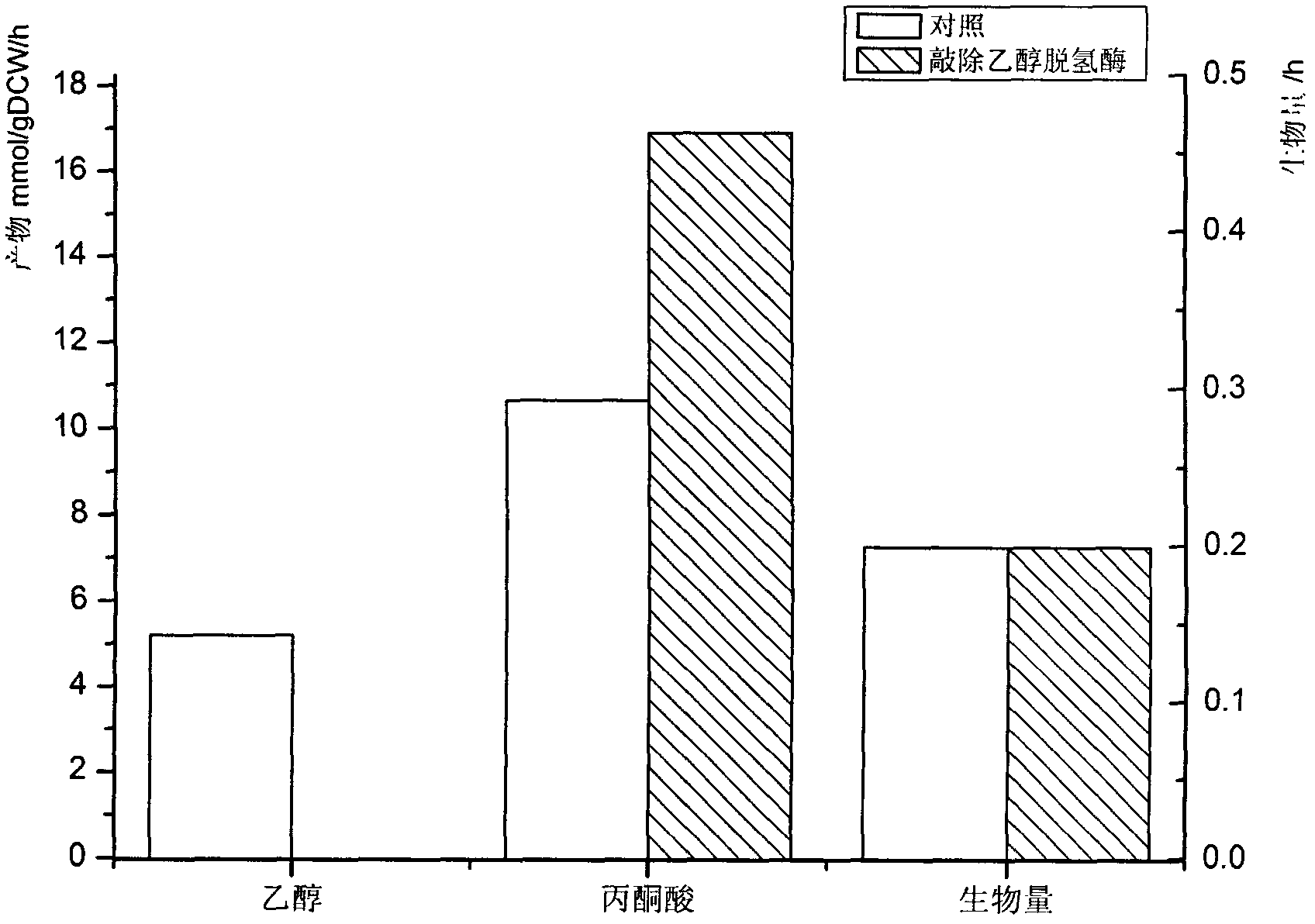
The invention discloses a construction method and an application technology of a Torulopsis glabrata genome-scale metabolism network model, belonging to the field of systems biology. The construction method of the model is an all-round semi-automatic construction method of combining comparative genomics annotation and bacteria-specific information of Torulopsis glabrata protein homology based on an automatic model. The invention provides a method for characterizing auxotroph characteristics in a Torulopsis glabrata metabolism network by adding necessary nutrient substances, lacked in bacteria, to a biomass equation. Alcohol dehydrogenase is knocked out by using single genes in the Torulopsis glabrata metabolism network model, and flow equilibrium analysis is applied, so that the output of pyruvic acid is increased. The construction method and the application technology of the Torulopsis glabrata genome-scale metabolism network model provide a high-efficient platform for fully understanding and reforming physiological and biochemical metabolisms of the Torulopsis glabrata.
Owner:JIANGNAN UNIV
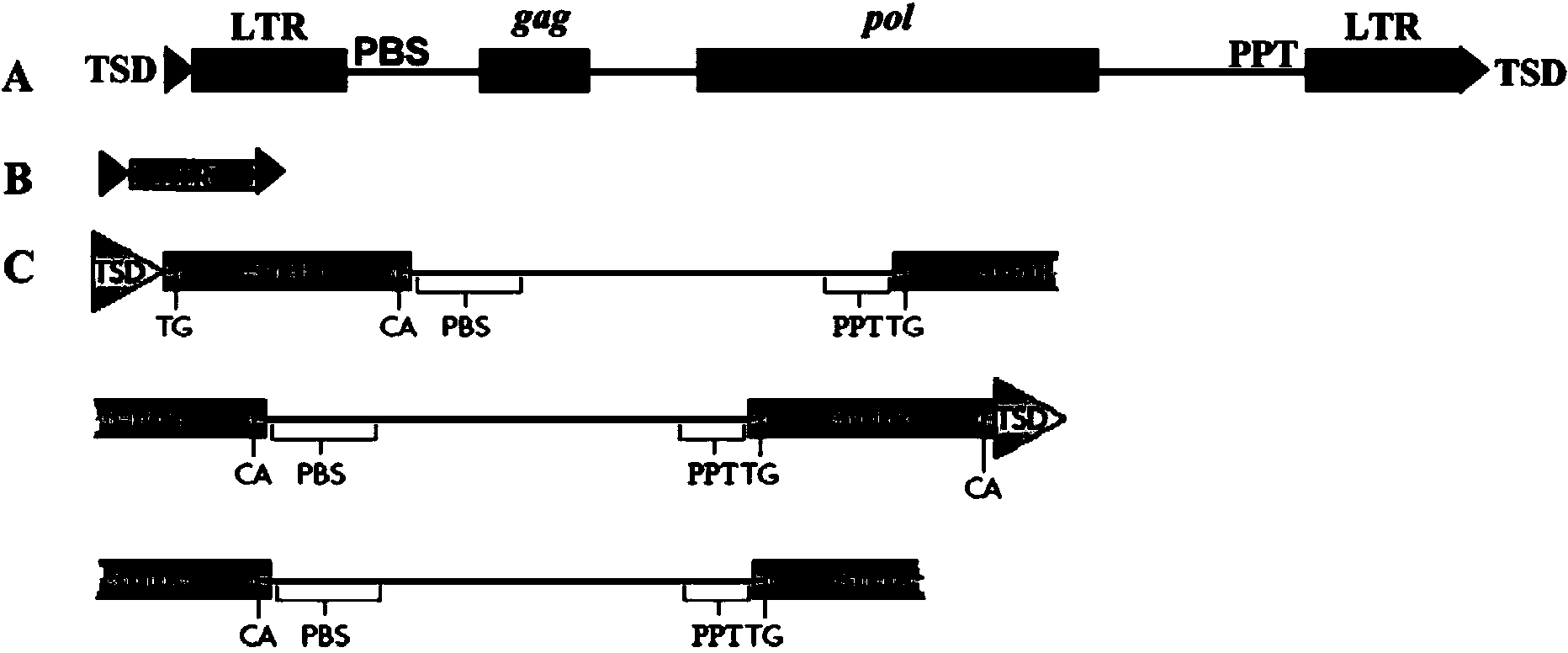
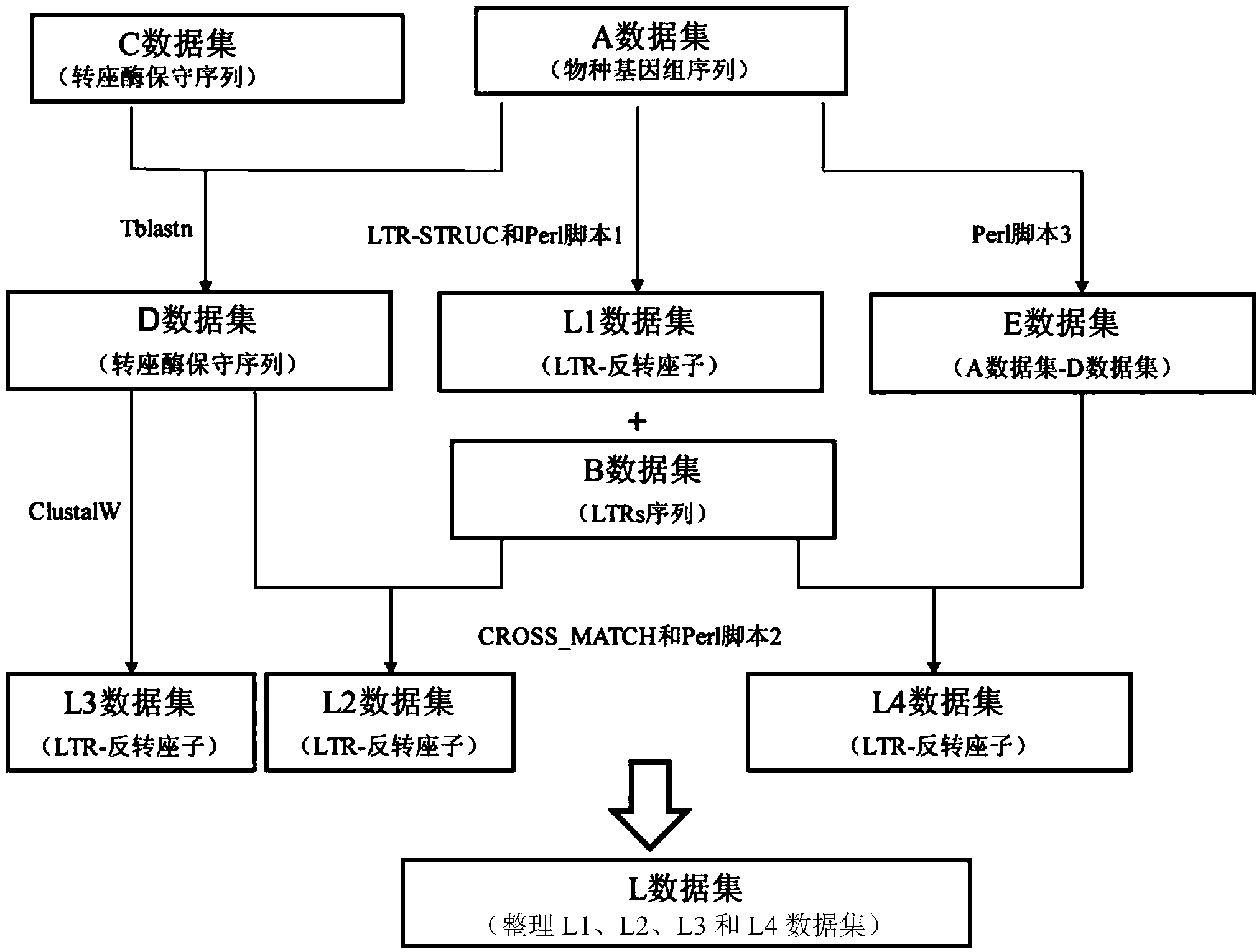
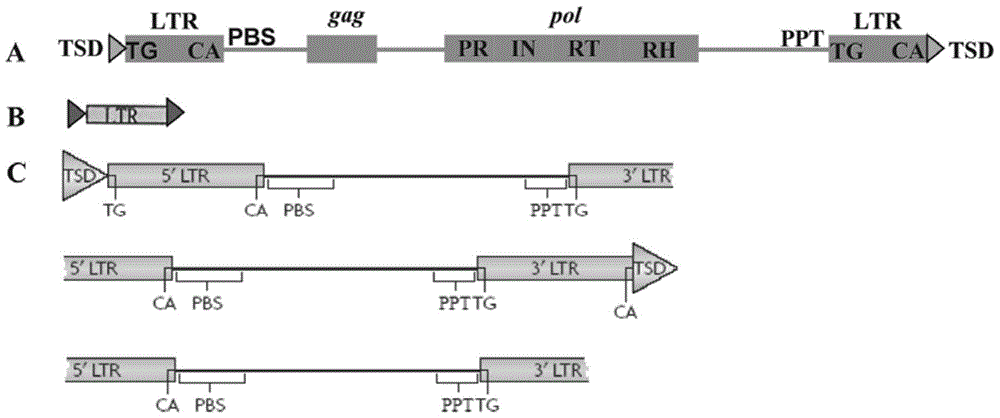
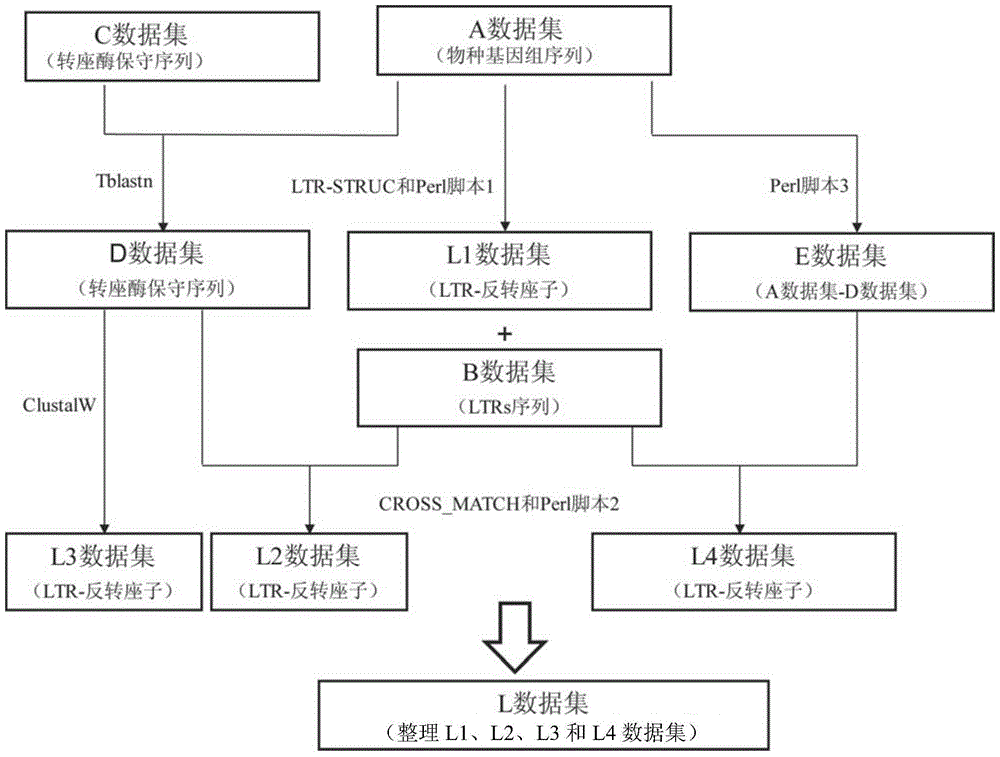
Method for batch inspection of plant genome LTR-retrotransposon
The invention discloses a method for batch inspection of plant genome LTR-retrotransposon. An LTR_STRUC program based on structural characteristics and searching from the beginning, a CROSS_MATCH program based on homology searching and a CLUSTALW comparison program based on sequence similarity are comprehensively applied in the method for the batch inspection of the plant genome LTR-retrotransposon, and Perl scripting language programming and other methods are combined. Experimental results show that the method for the batch inspection of the plant genome LTR-retrotransposon is relatively systematic, the effect for detecting the direct repeat of the inserting sites of the plant genome LTR-retrotransposon is good, speed is high, and process operation is easy to achieve. According to the method, the frequently-used software for detecting the plant genome LTR-retrotransposon and the Perl scripting language programming are combined, and certain defects of the frequently-used software are made up. The method can play an important role in genome annotation and the batch inspection of the plant genome LTR-retrotransposon.
Owner:JIANGSU ACAD OF AGRI SCI
Reverse engineering genome-scale metabolic network reconstructions for organisms with incomplete genome annotation and developing constraints using proton flux states and numerically-determined sub-systems
InactiveUS8311790B2Analogue computers for chemical processesSystems biologyInformation repositoryMetabolic network
A genome-scale metabolic network reconstruction for Clostridium acetobutylicum (ATCC 824) was created using a new semi-automated reverse engineering algorithm. This invention includes algorithms and software that can reconstruct genome-scale metabolic networks for cell-types available through the Kyoto Encyclopedia of Genes and Genomes. This method can also be used to complete partial metabolic networks and cell signaling networks where adequate starting information base is available. The software may use a semi-automated approach which uses a priori knowledge of the cell-type from the user. Upon completion, the program output is a genome-scale stoichiometric matrix capable of cell growth in silico. The invention also includes methods for developing flux constraints and reducing the number of possible solutions to an under-determined system by applying specific proton flux states and identifying numerically-determined sub-systems. Although the model-building and analysis tools described in this invention were initially applied to C. acetobutylicum, the novel algorithms and software can be applied universally.
Owner:UNIVERSITY OF DELAWARE
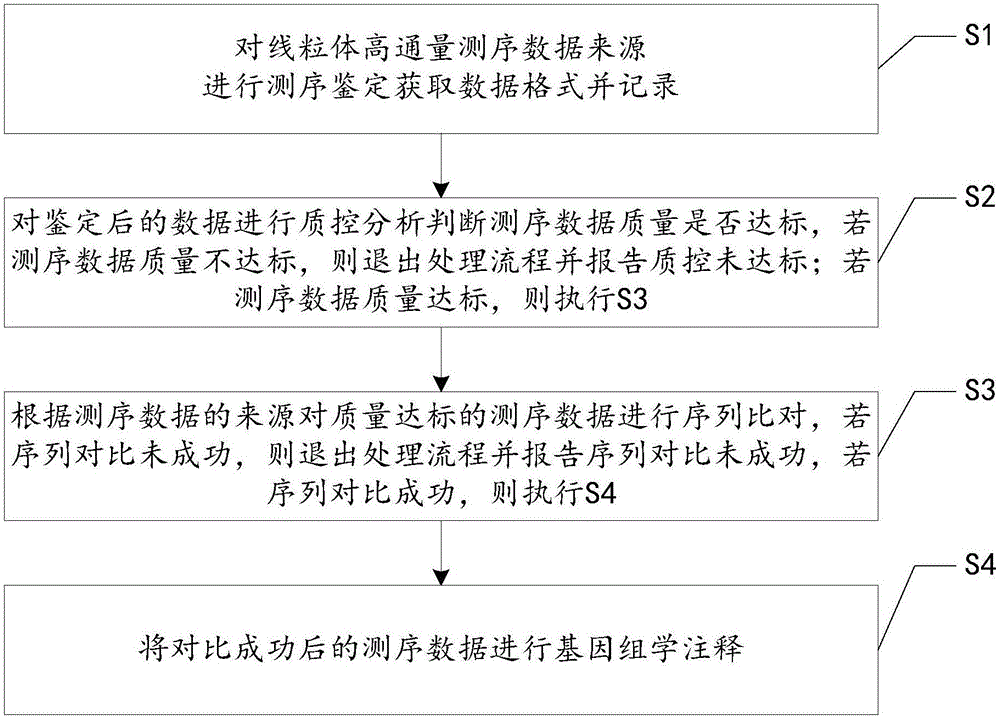
Processing method and system for mitochondrial high-throughput sequencing data
InactiveCN106650308AConvenient for clinical operationFacilitate scientific researchProteomicsGenomicsGenomicsThe Internet
The invention relates to the technical field of bioinformation, in particular to a processing method and system for mitochondrial high-throughput sequencing data. The method includes the steps that sequencing authentication is conducted on the source of the mitochondrial high-throughput sequencing data to acquire the data format, and then the data format is recorded; quality control analysis is conducted on the authenticated data to judge whether the quality of the sequencing data reaches the standard or not, and if the quality of the sequencing data does not reach the standard, the processing process is quitted and it is reported that quality control does not reach the standard; if the quality of the sequencing data reaches the standard, sequence alignment is conducted on the sequencing data with the quality reaching the standard according to the source of the sequencing data; if sequence alignment does not succeed, the processing process is quitted and it is reported that sequence alignment does not succeed, and if sequence alignment succeeds, genomic annotation is conducted on the sequencing data obtained after successful sequence alignment. By means of the processing method and system, high-automation batched analysis and processing can be conducted on the mitochondrial gene detection data without Internet connection, and gene variations can be displayed in a classified mode according to clinical detection and experimental and scientific research, so that convenience is brought to clinical application and scientific research.
Owner:神州医学数据科技(北京)有限公司
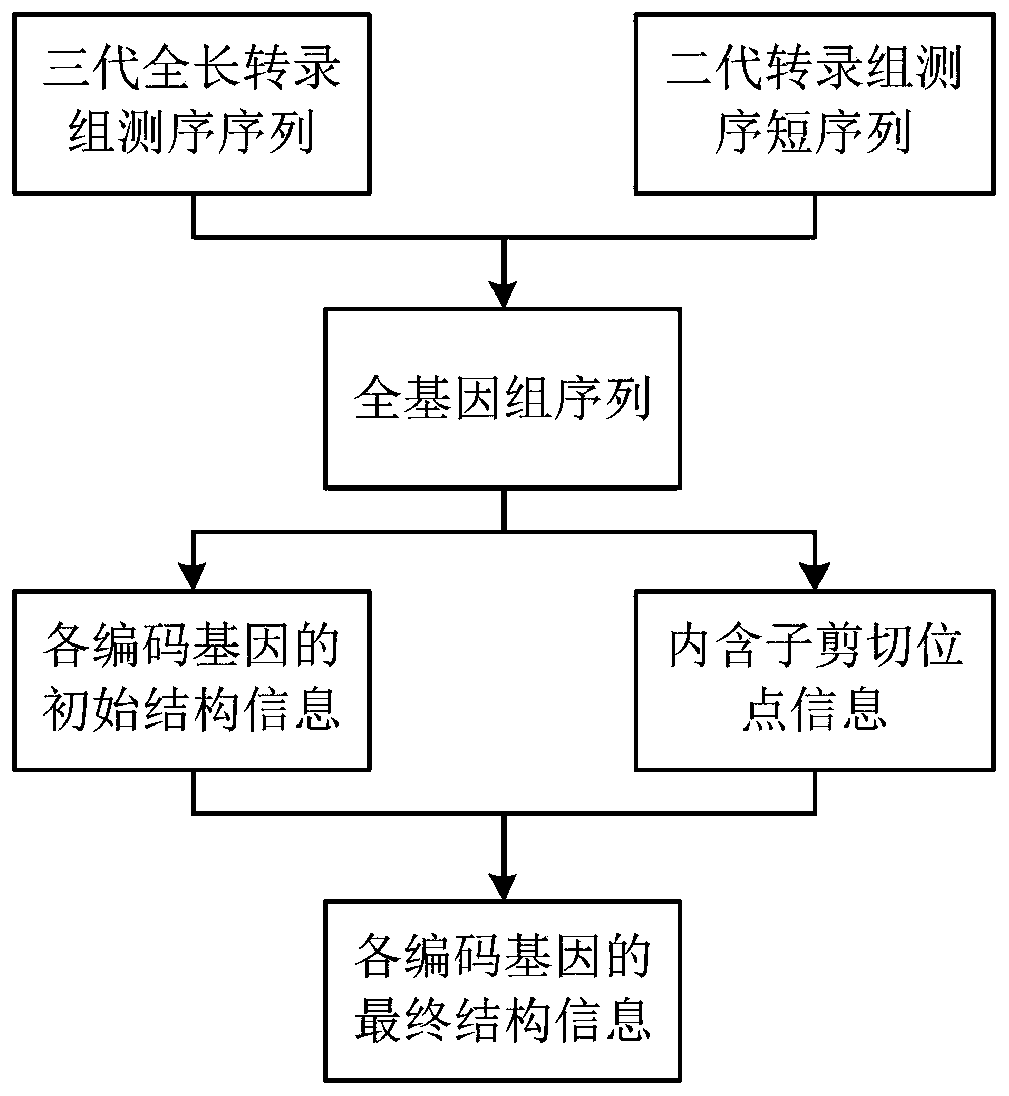
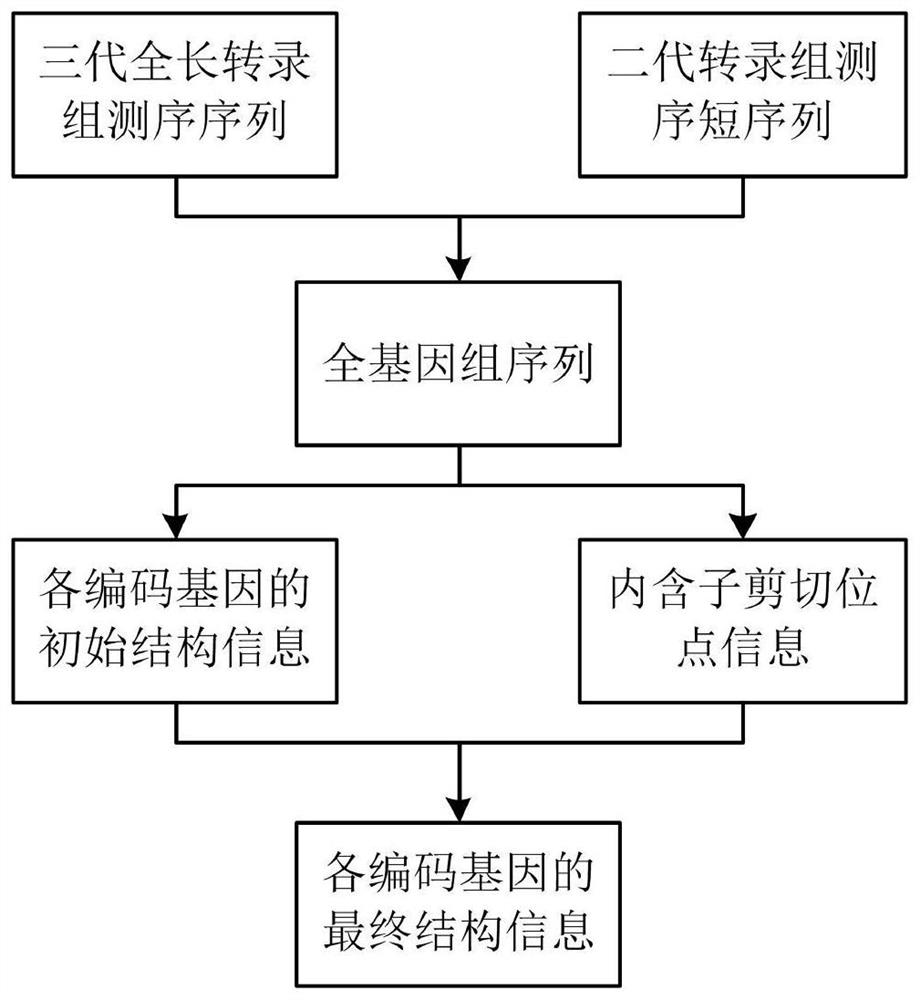
Genome annotation method utilizing second-generation and third-generation transcriptome sequencing data
ActiveCN111312331AImprove accuracyImprove annotation efficiencyProteomicsGenomicsTranscriptome SequencingGenomic annotation
The invention relates to the technical field of genome annotation, and provides a genome annotation method utilizing second-generation and third-generation transcriptome sequencing data. The method comprises the following steps: 1, comparing a third-generation full-length transcriptome sequencing sequence to a target genome to obtain initial structure information of each encoding gene; 2, comparing the second-generation transcriptome sequencing short sequence to a target genome, and extracting intron cleavage site information from a comparison file; and 3, obtaining final structure informationof each coding gene by combining the initial structure information of each coding gene and the intron cleavage site information. According to the method, a full-length transcript sequence can be obtained without splicing a third-generation transcriptome and second-generation transcriptome sequencing data can provide a large amount of intron cleavage site evidences can also be fully considered, and the accuracy and efficiency of genome annotation are greatly improved.
Owner:三亚博瑞源科技有限公司
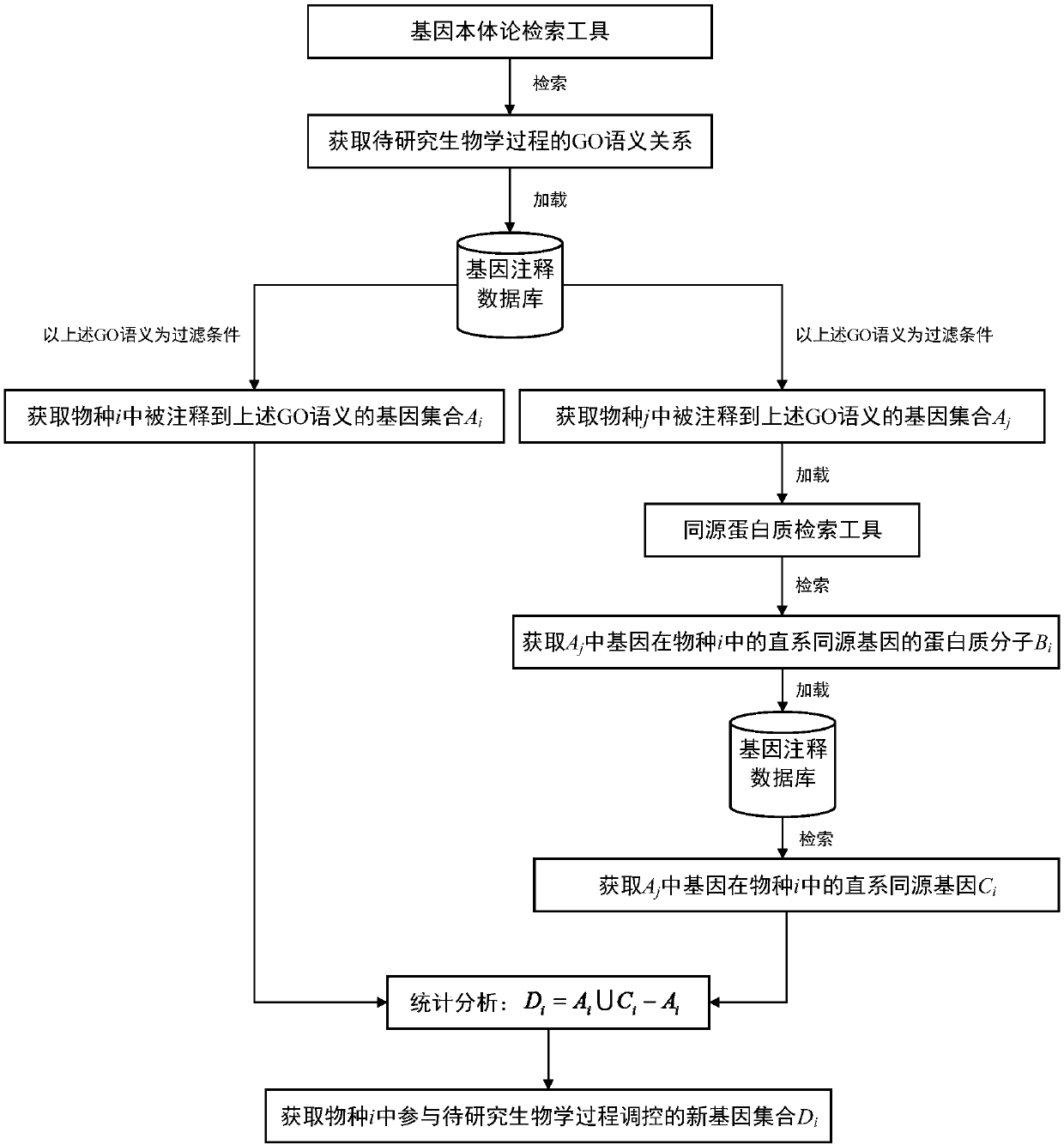
Method for screening regulation new gene participating in biological process
The invention discloses a method for screening a regulation new gene participating in the biological process. A biologic informatics method is used for searching gene ontology for a tool, and the semantic relation in the specific biological process is screened out; in a genome annotation database of different species, gene set information in different species annotated to the semantic is screenedout; homology analysis is performed on the gene set information of the specific species, an orthologous gene in the species to be researched is screened out; comparison and analysis are performed on the gene and the reference gene participating in the biologic process in the species to be researched, and the regulation new gene participating in the biologic process in the specific species is screened out. On the basis that the specific biologic process has the advantage of high conservative property between different species, a method for screening the regulating new gene participating in thebiologic process is built, the supporting effect is supplied for further reconstructing a systematic gene regulation network, and the method is of a great significance in early disease diagnosis, individual treatment and medicine development.
Owner:DALIAN MARITIME UNIVERSITY
Construction system of novel transposon mutant strain library, novel transposon mutant library and application
ActiveCN114317584AReduce capacityIncrease diversityBacteriaMicroorganism based processesBiotechnologyPseudomona aeruginosa
The invention discloses a construction system of a novel transposon mutation library, plasmids can be transferred into a target strain through chemical conversion and electric conversion methods, then expression of transposase is induced, and the transposon mutation library almost covering the whole genome range can be obtained only through a mutant strain containing transposon. The construction system of the novel transposon library can not only increase the diversity of transposon mutation, but also facilitate the determination of the insertion position of a strain, and facilitate the research on genome annotation, functional analysis and drug resistance mechanism of pseudomonas aeruginosa.
Owner:HAINAN MEDICAL COLLEGE +1
Method for efficiently purifying and identifying cyclic RNA based on second-generation Illumina sequencing platform
InactiveCN110283895AEfficient identificationImprove removal efficiencyMicrobiological testing/measurementData setRNA Sequence
The invention discloses a method for efficiently purifying and identifying cyclic RNA based on a second-generation Illumina sequencing platform and belongs to the field of molecular biology. By use of a cyclic RNA sequencing library obtained by purification through the method, the removing efficiency of unrelated linear RNA can be greatly improved, and high-purity cyclic RNA is obtained by enriching. A virtual data set of the cyclic RNA is constructed on the basis of genome annotation. By searching and comparing reads of sequencing data, the types of the identified cyclic RNA are far higher than that of a conventional method. The method has important significance for researching an internal structure of the cyclic RNA and revealing the functions of the cyclic RNA.
Owner:FUJIAN AGRI & FORESTRY UNIV
Systems and methods for genomic annotation and distributed variant interpretation
A computer-based genomic annotation system, including a database configured to store genomic data, non-transitory memory configured to store instructions, and at least one processor coupled with the memory, the processor configured to implement the instructions in order to implement an annotation pipeline and at least one module filtering or analysis of the genomic data
Owner:THE SCRIPPS RES INST
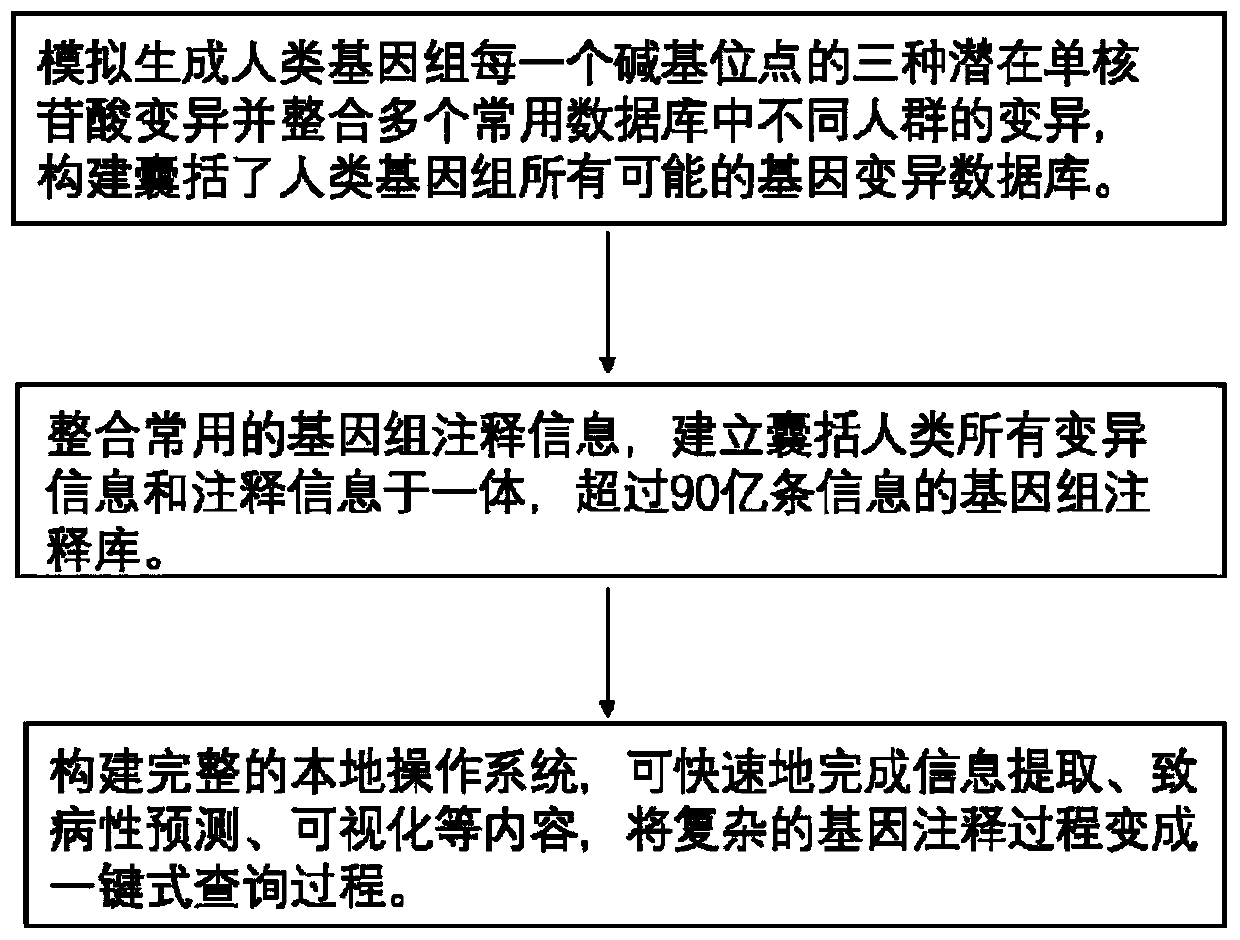
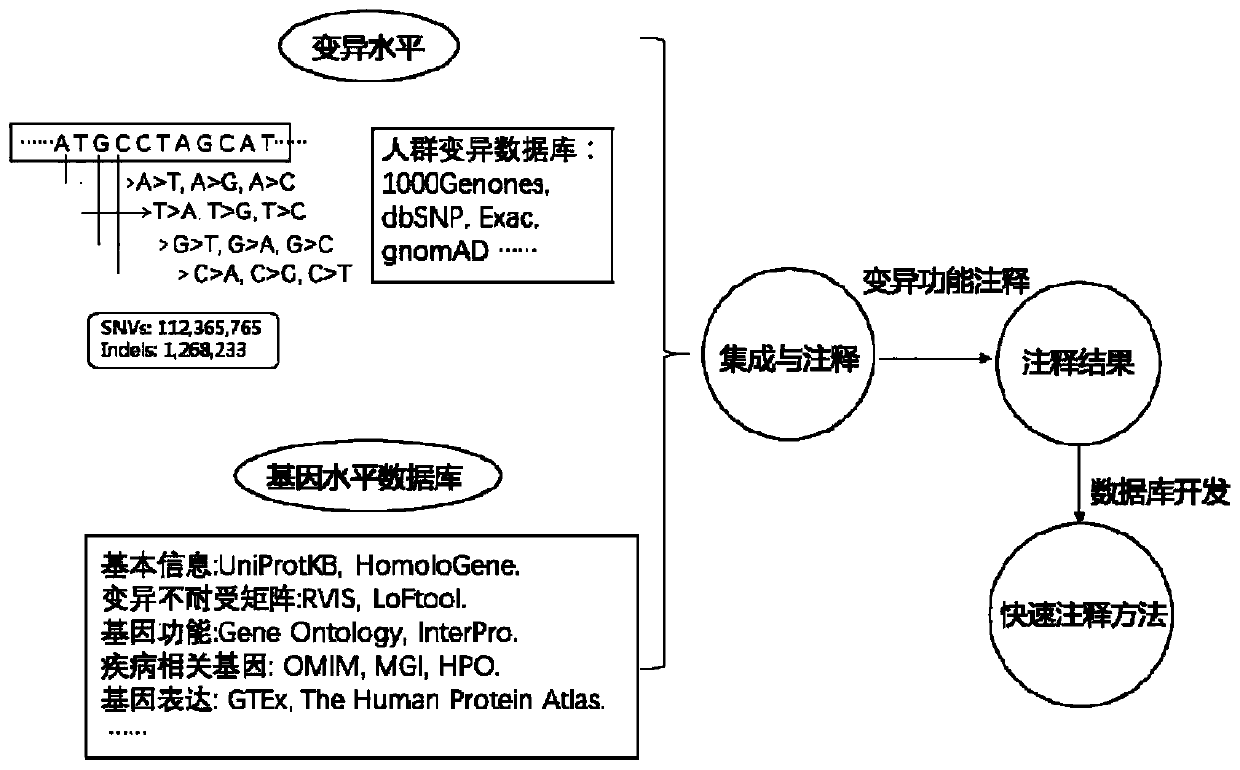
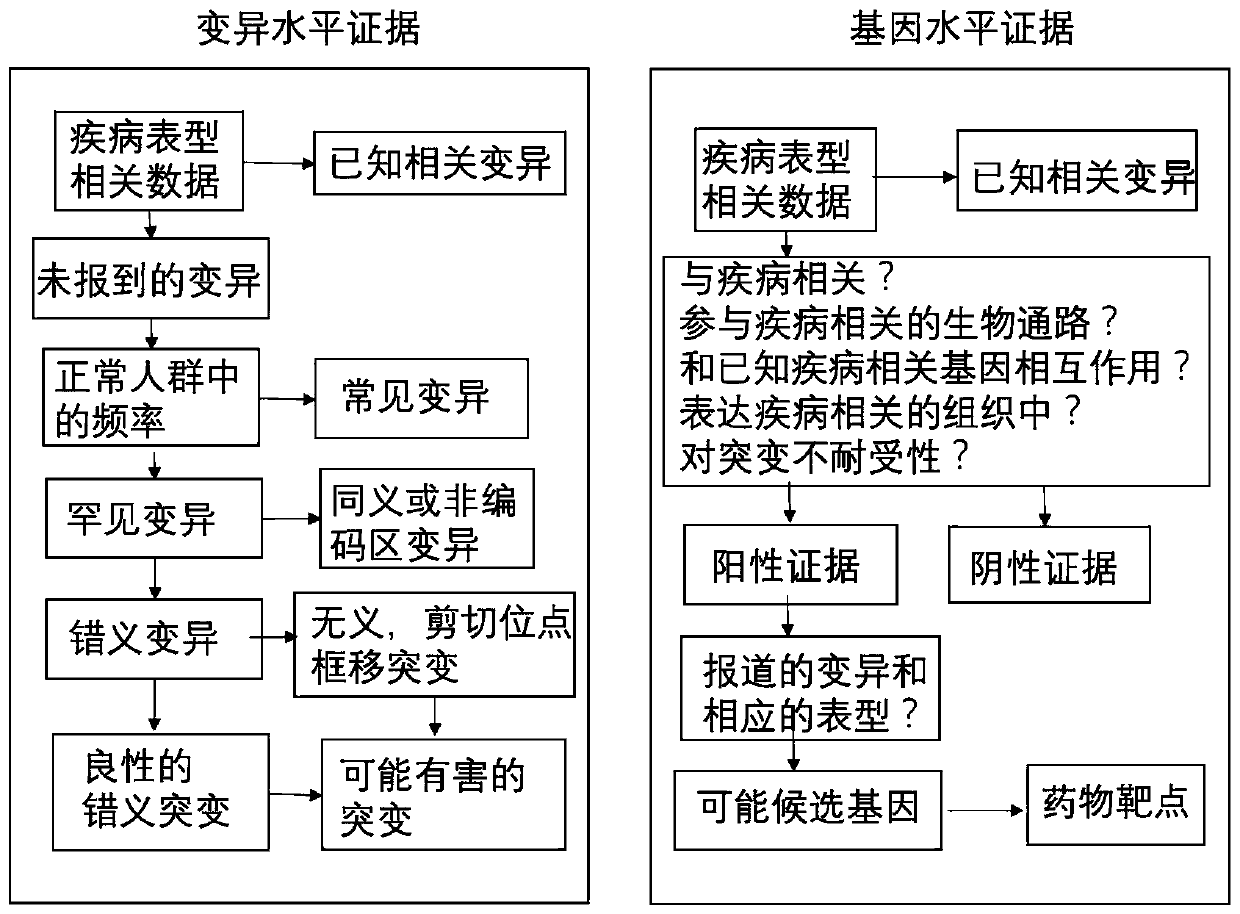
Method for quickly annotating gene mutations of human beings
The invention is applicable to the technical field of medical genetics, and provides a novel method for quickly annotating genes and mutations by constructing an information query database with complete annotations. The method comprises the following steps: (1) simulating to generate three potential mononucleotide variations of each base site of a human genome, and integrating the variations of different populations in a plurality of common databases to construct a database including all possible gene variations of the human genome; (2) further integrating common genome annotation informationon the basis, and establishing a genome annotation library including all variation information and annotation information of human beings into a whole and more than 9 billion pieces of information; and (3) quickly completing information extraction, pathogenicity prediction, visualization and other content in a query mode according to the database. According to the method, a complex gene annotationprocess is changed into a one-stop query process, so that the problems of non-uniform formats, complicated operation and low efficiency caused by matching different gene variations in multiple databases one by one are avoided.
Owner:XIANGYA HOSPITAL CENT SOUTH UNIV
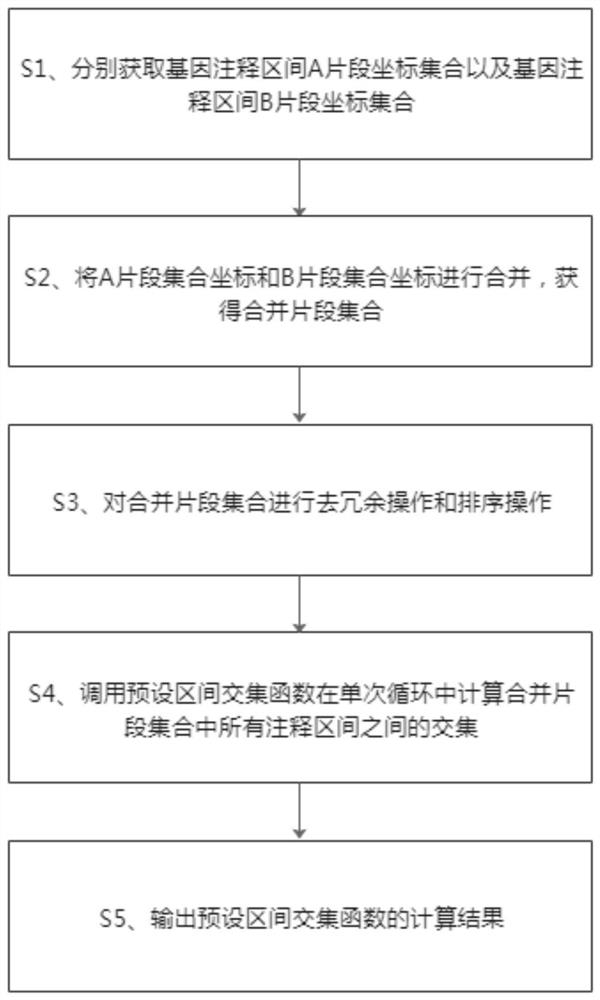
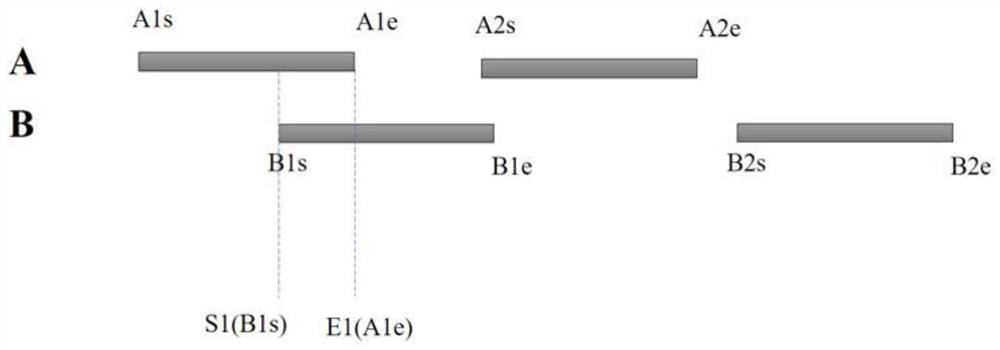
Method and system for quickly comparing whole genome annotation intervals
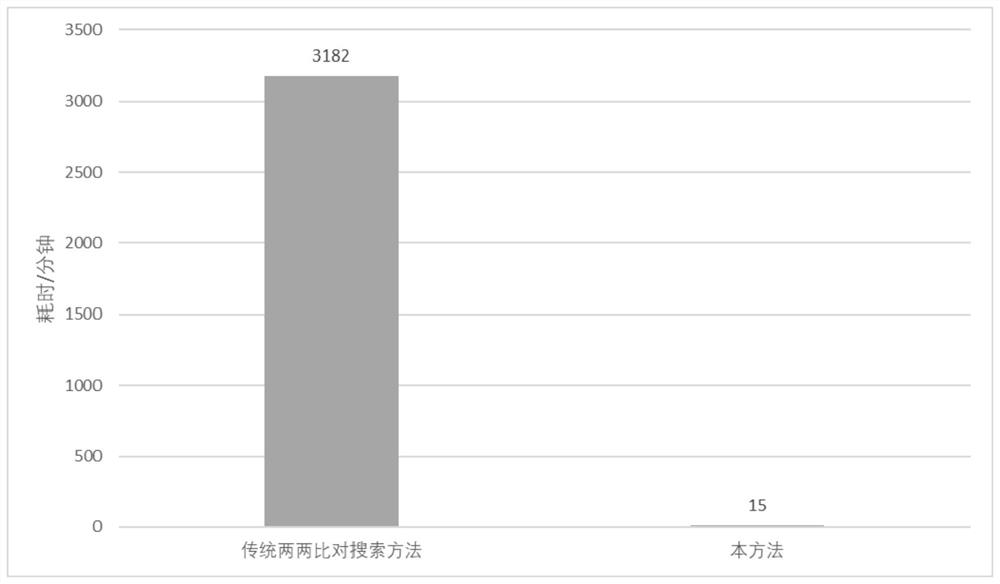
The invention provides a method and a system for quickly comparing whole genome annotation intervals. The method comprises the following steps: S1, respectively obtaining a gene annotation interval A fragment coordinate set and a gene annotation interval B fragment coordinate set; s2, combining the coordinates of the fragment set A and the coordinates of the fragment set B to obtain a combined fragment set; s3, performing redundancy elimination operation and sorting operation on the combined fragment set; s4, calling a preset interval intersection function to calculate the intersection of all the annotation intervals in the merged fragment set in a single cycle; and S5, outputting a calculation result of the preset interval intersection function. According to the method, the intersection of different types of gene annotation intervals can be rapidly calculated, and compared with a traditional comparison method, the method is simple in implementation logic, small in calculation amount, accurate in judgment and beneficial to improvement of comparison efficiency.
Owner:HAINAN UNIVERSITY
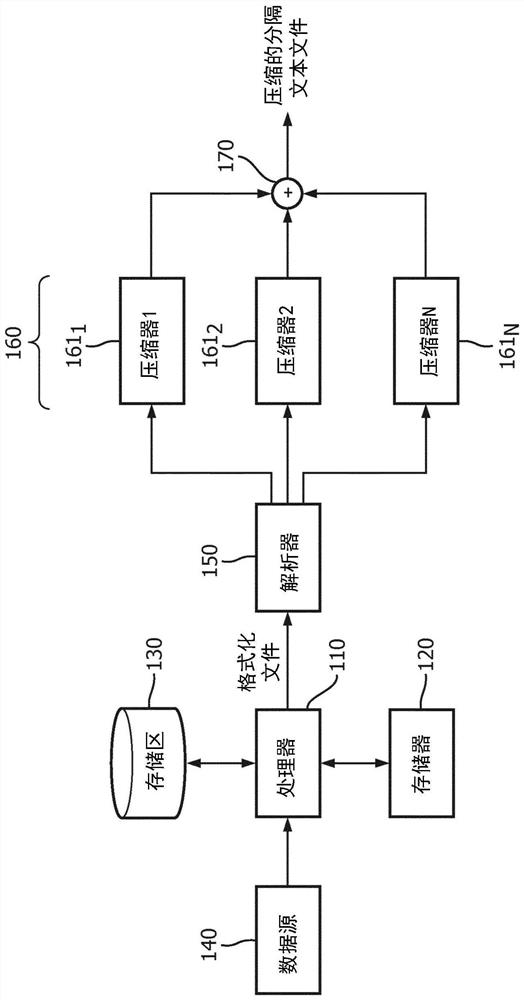
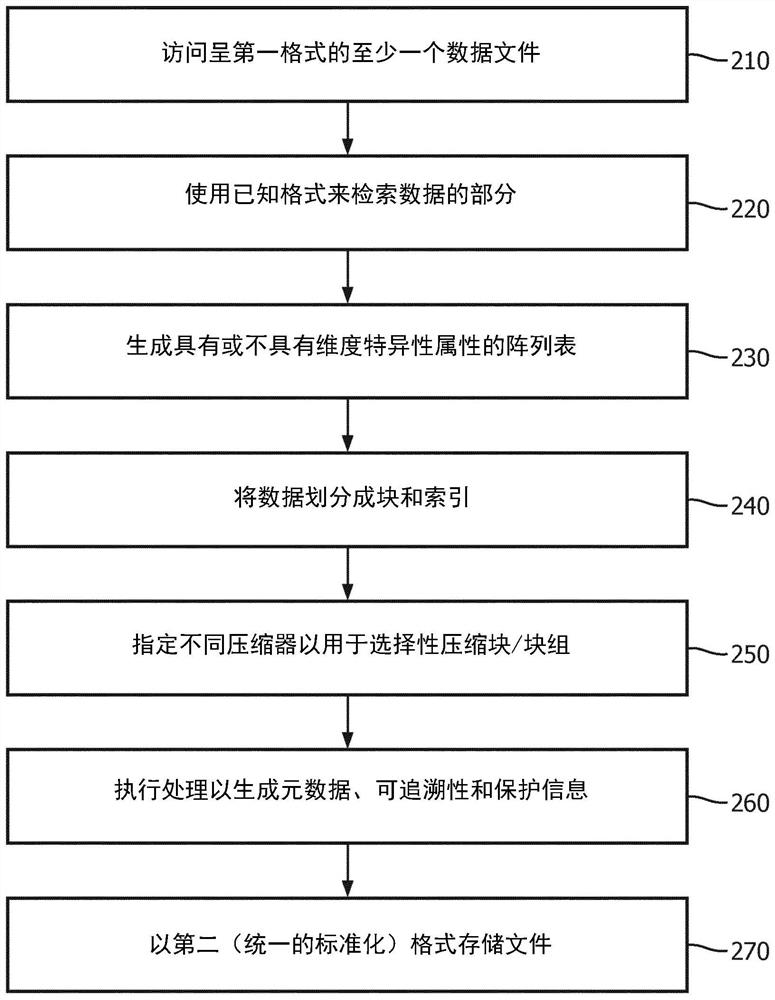
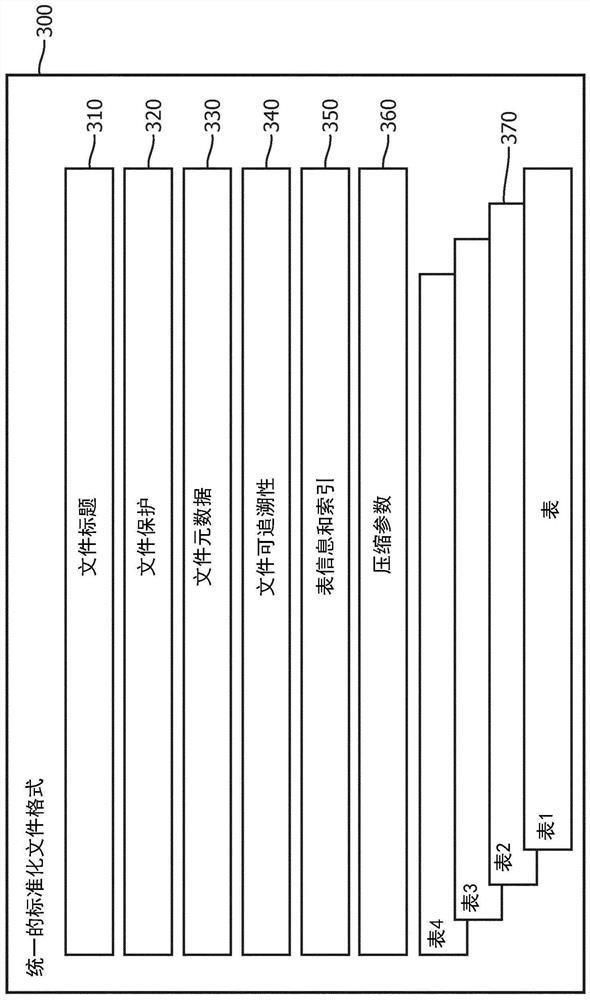
Systems and methods for efficient compression, representation and decompression of a variable table data
A method for controlling data compression includes accessing genomic annotation data in one of a plurality of first file formats; extracting attributes from the genome annotation data; dividing the genome annotation data into blocks; and processing the extracted attributes and blocks into related information. The method further includes selecting a different compressor for the attribute and the block identified in the correlation information; and generating a file in a second file format, the file in the second file format including the related information and information indicating the different compressors for the attribute and the chunk indicated in the related information. The information indicating the different compressor is processed into the second file format to allow for selective decompression of the attributes and the chunks indicated in related information.
Owner:KONINKLJIJKE PHILIPS NV
Method for assembling and annotating Guide black fur sheep genome based on three-generation PacBio and Hi-C technologies
PendingCN113005189AQuality improvementImprove integrityMicrobiological testing/measurementProteomicsBiotechnologyGenetics
The invention belongs to the technical field of biological information, and particularly relates to a method for assembling and annotating a Guide black fur sheep genome based on three-generation PacBio and Hi-C technologies. The method comprises the following steps: (1) collecting blood and tissue samples of Gueide black fur sheep; (2) constructing a genome library and a transcriptome library; (3) performing genome size and heterozygosis rate evaluation; (4) carrying out genome assembly, and carrying out error correction and evaluation by utilizing a transcription sequencing result; (5) carrying out Hi-C auxiliary assembly, error correction and evaluation; and (6) carrying out genome annotation and evaluation. The chromosome-level high-quality genome of the Guide black fur sheep is assembled, a foundation is laid for further research on wool pigment deposition and high-cold low-oxygen adaptive molecular mechanism of the Guide black fur sheep, and data support is provided for work of protection and utilization of excellent genetic resources, molecular breeding, genetic improvement and the like of the Guide black fur sheep.
Owner:LANZHOU INST OF ANIMAL SCI & VETERINARY PHARMA OF CAAS
A method for accurate prediction of BSA-seq candidate genes from low-density SNP genomic regions
ActiveCN109360606BMake up for false positivesMicrobiological testing/measurementProteomicsConfidence intervalCandidate Gene Association Study
The invention relates to the genome sequencing technology field and especially relates to a method for accurately predicting a BSA-seq candidate gene in a low-density SNP genomic region. BSA-seq has alow-density SNP region near a candidate interval. Through comparing SNPs between two parents, an SNP list is strictly filtered to find a low-density region, and then, a corresponding candidate interval when a confidence interval is 95% and a low-density candidate interval are used, and a genome annotation website is used to annotate the genes in a candidate region. A candidate region variation point function is annotated so as to obtain a gene with functional variation such as frameshift variation and the like, and the gene is determined to be a candidate gene. The method can be used to makeup for the false positive of the candidate region due to the region with a small difference in a genome, and the real candidate interval is acquired.
Owner:广西壮族自治区农业科学院水稻研究所
Systems and methods for genomic annotation and distributed variant interpretation
A computer-based genomic annotation system, including a database configured to store genomic data, non-transitory memory configured to store instructions, and at least one processor coupled with the memory, the processor configured to implement the instructions in order to implement an annotation pipeline and at least one module filtering or analysis of the genomic data.
Owner:THE SCRIPPS RES INST
Systems and methods for genomic annotation and distributed variant interpretation
Owner:THE SCRIPPS RES INST
Method for screening genes related to synthesis of target compound and application
PendingCN113337589AImprove accuracyAchieve high quality and high yieldMicrobiological testing/measurementTranscriptome SequencingGenomic annotation
The invention discloses a method for screening genes related to synthesis of a target compound and application. The method in the invention comprises the following steps of: taking a third-generation transcript as a reference transcript, and comparing second-generation sequencing data to the transcript to obtain the abundance of each transcript; and quantifying the second-generation assembled transcript by using RSEM software, and carrying out differential expression analysis between genes to obtain a key gene in the terpenoid synthesis process. Compared with a second-generation transcriptome sequencing technology and a third-generation full-length transcriptome sequencing technology which are independently used, the invention has the advantages that: the advantage that the full-length transcriptome sequence can be obtained without splicing the third-generation transcriptome can be sufficiently utilized, so that the accuracy is high; the advantage of quantitative expression detection of genes by second-generation transcriptome sequencing data can be fully considered; the accuracy of genome annotation is greatly improved; and related genes in a synthetic route of the target compound are obtained through screening. According to the method, four genes related to cinnamomum burmannii monoterpenoid synthetase are obtained for the first time.
Owner:SOUTH CHINA UNIV OF TECH
A Genome Annotation Method Using Second- and Third-Generation Transcriptome Sequencing Data
ActiveCN111312331BImprove accuracyImprove annotation efficiencyProteomicsGenomicsTranscriptome SequencingGenomic annotation
The invention relates to the technical field of genome annotation, and provides a genome annotation method using second-generation and third-generation transcriptome sequencing data. The method of the present invention includes the following steps: Step 1: align the three-generation full-length transcriptome sequencing sequence to the target genome to obtain the initial structural information of each coding gene; Step 2: align the second-generation transcriptome sequencing short sequence to the target genome Genome, extract intron splicing site information from the comparison file; step 3: combine the initial structure information of each coding gene and intron splicing site information to obtain the final structure information of each coding gene. The present invention can not only make full use of the advantage of high accuracy that the full-length transcript sequence can be obtained without splicing the third-generation transcriptome, but also fully consider the advantage that the second-generation transcriptome sequencing data can provide a large number of intron splicing site evidences , greatly improving the accuracy and efficiency of genome annotation.
Owner:三亚博瑞源科技有限公司
A method for batch detection of ltr-retrotransposons in plant genomes
ActiveCN103824000BGood effectHigh speedSpecial data processing applicationsDirect repeatStructure based
The invention discloses a method for batch inspection of plant genome LTR-retrotransposon. An LTR_STRUC program based on structural characteristics and searching from the beginning, a CROSS_MATCH program based on homology searching and a CLUSTALW comparison program based on sequence similarity are comprehensively applied in the method for the batch inspection of the plant genome LTR-retrotransposon, and Perl scripting language programming and other methods are combined. Experimental results show that the method for the batch inspection of the plant genome LTR-retrotransposon is relatively systematic, the effect for detecting the direct repeat of the inserting sites of the plant genome LTR-retrotransposon is good, speed is high, and process operation is easy to achieve. According to the method, the frequently-used software for detecting the plant genome LTR-retrotransposon and the Perl scripting language programming are combined, and certain defects of the frequently-used software are made up. The method can play an important role in genome annotation and the batch inspection of the plant genome LTR-retrotransposon.
Owner:JIANGSU ACAD OF AGRI SCI
Systems and methods for genomic annotation and distributed variant interpretation
A computer-based genomic annotation system, including a database configured to store genomic data, non-transitory memory configured to store instructions, and at least one processor coupled with the memory, the processor configured to implement the instructions in order to implement an annotation pipeline and at least one module filtering or analysis of the genomic data.
Owner:THE SCRIPPS RES INST
A promoter query method, system and device for a species genome
ActiveCN113284559BImprove accuracyAdapt to the needs of transformation carriersDigital data information retrievalSequence analysisGenomic annotationPromoter
The invention relates to a promoter query method, system and equipment of a species genome, the method comprises obtaining the genome file, genome annotation file and fastq file of the species; according to the genome annotation file, sorting all the genes in the genome file to obtain Species promoter files; use hisat2 or Trinity software to convert fastq files into FPKM files containing highly expressed genes; use steps S1 to S3 to obtain species promoter files and FPKM files of several species, and pass the species promoter files of several species and FPKM files to build a promoter database. The promoter query method of the genome of this species can query the promoters of the required genes in the promoter database, does not limit the query of gene promoters of any species, does not require additional auxiliary tools, and the accuracy of the promoters obtained from the query is high .
Owner:JINAN UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com