Construction and application technology of Torulopsis glabrata genome metabolism model
A technology of T. glabrata and genome, applied in the field of systems biology, can solve the problems of uneven model quality and so on
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
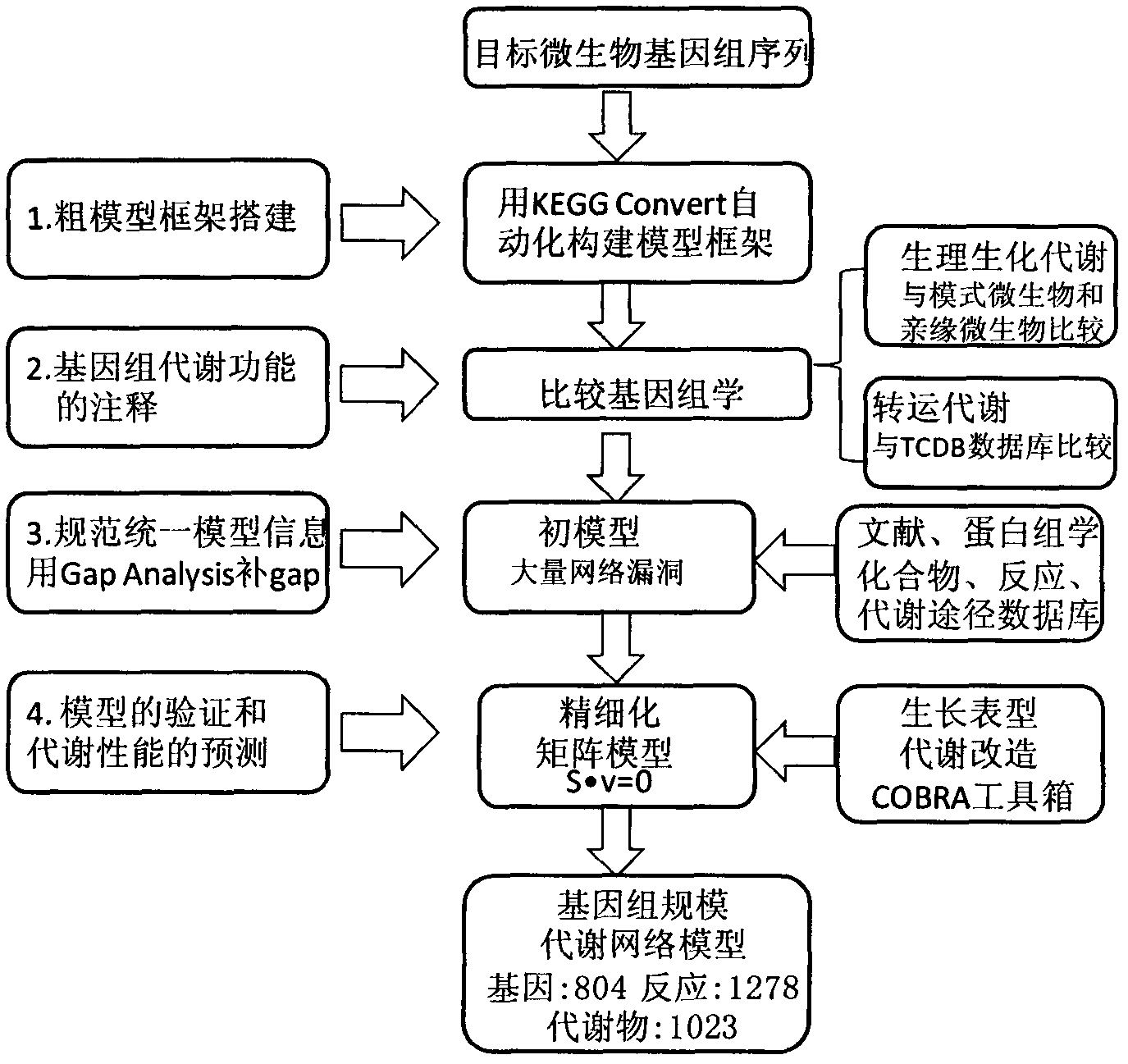
[0011] Example 1 A model framework is built using KEGG converter.
[0012] Download the KGML format file of the metabolic pathway of Torulopsis glabrata from the KEGG database, and use KEGG converter software to integrate all metabolic pathways into a metabolic network model in Systems Biology Markup Language (SBML) format.
Embodiment 2
[0013] Example 2 Annotation of the genome sequence of Toruula glabrata based on protein sequence homology
[0014] The comparison strains were selected among microorganisms for which genome-scale metabolic network models had been constructed. First, starting from the phylogenetic tree, select microorganisms that belong to the same family with close kinship. Saccharomyces cerevisiae and Pichia pastoris were selected here as close relatives of Toruula glabrata. To ensure the comprehensiveness of gene annotation, model microorganisms such as Aspergillus niger, Escherichia coli, and Bacillus subtilis were also used as comparison strains.
[0015] Download the FASTA format of the protein sequences of the above microorganisms from UniprotKB, and perform BLASTp with the protein sequences of Torulopsis glabrata. The comparison results meet the similarity greater than 35% and the E value is less than 10 -30 , and the ratio of similar length to any sequence length is greater than 70%...
Embodiment 3
[0018] Example 3 Metabolic network model combined with bibliographic studies of Toruula glabrata
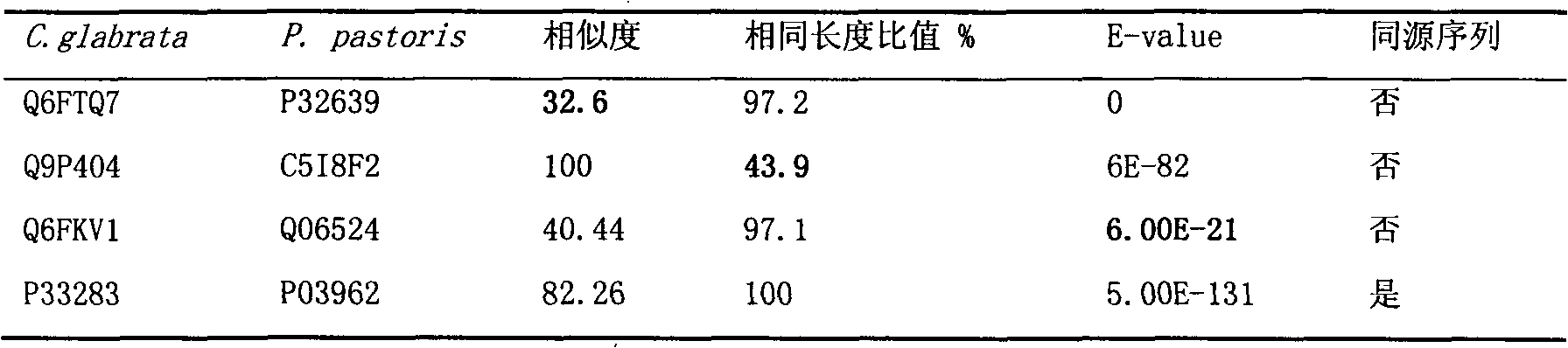
[0019] Using (Candida glabrata or Torulopsis glabrata) and (metabolism or physiology) as keywords, the literature related to the physiological and biochemical metabolism of Torulopsis glabrata was excavated in PubMed and Web of Science. According to literature reports, 254 reactions were confirmed to exist in Torulopsis glabrata, and 82 putative proteins were functionally annotated (this annotation does not coincide with the gene annotations of KEGG and comparative genomics). Partial annotation information is shown in Table 2.
[0020] Table 2 Partial information of gene annotation based on bibliographic omics
[0021]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com