Method for accurately predicting BSA-seq candidate gene in low-density SNP genomic region
A candidate gene and accurate prediction technology, applied in genomics, biochemical equipment and methods, proteomics, etc., can solve problems such as low accuracy, false positives in candidate regions, and low precision
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
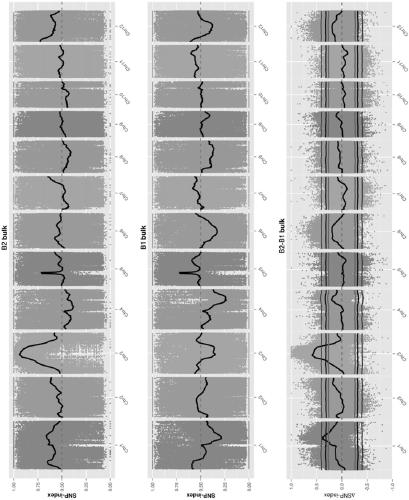
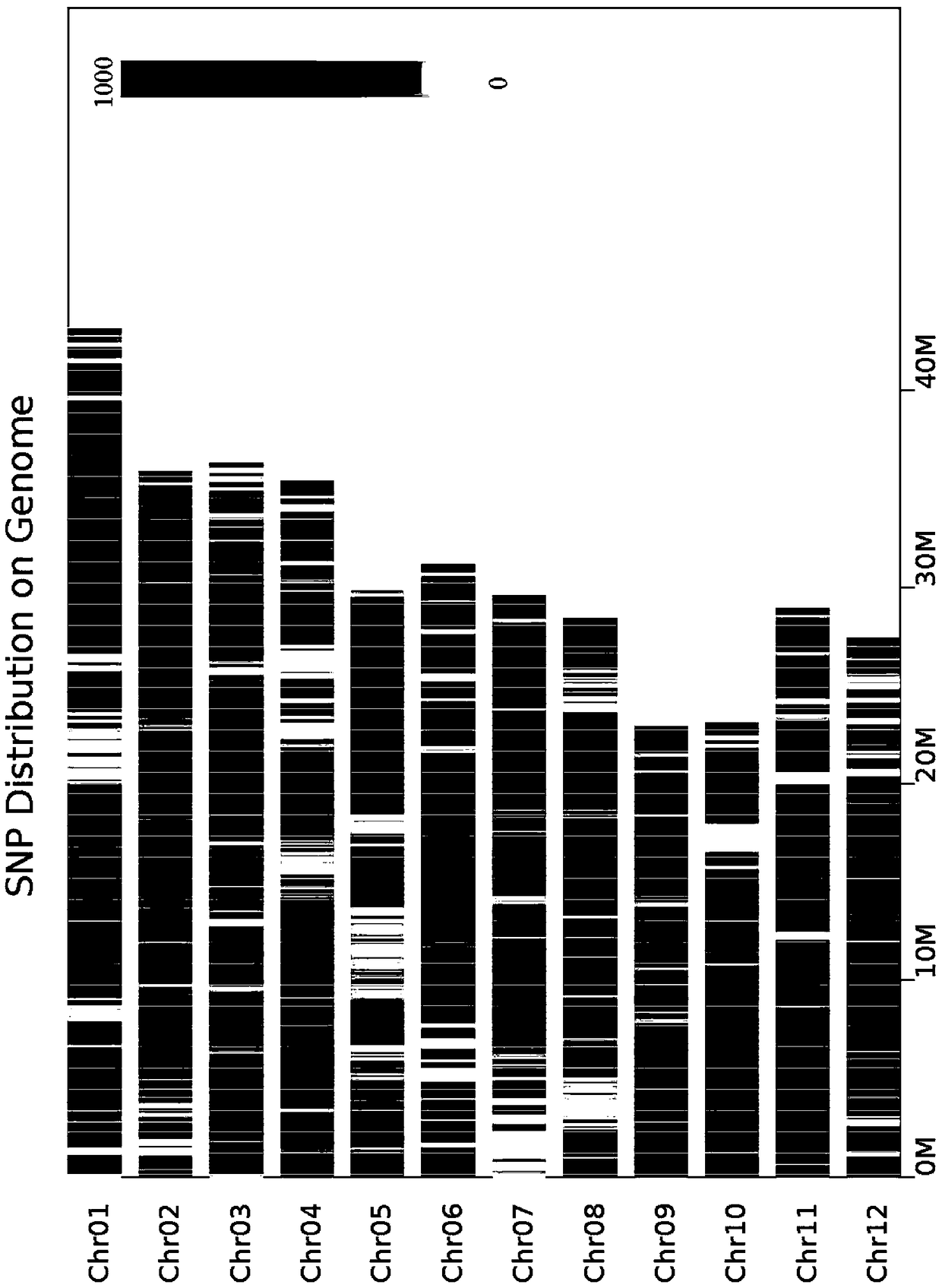
[0030] This embodiment provides a method for accurately predicting BSA-seq candidate genes in a low-density SNP genomic region, and the steps of the method are as follows:
[0031] (1) Pool mixing: select the parent "Huanghuazhan × Donglan Momi" with significant differences in the target traits to construct a segregation population, and then select 30-50 individual plants with extreme phenotypes of the target traits from the segregation population, and mix them into two DNAs Pool (DNA pools) for sequencing;
[0032] (2) extracting DNA: using the CTAB method to extract plant genomic DNA;
[0033] (3) Sequencing: After the genomic DNA sample obtained in step (2) is qualified, the DNA is randomly broken into 350bp fragments by ultrasonic crushing, and the DNA fragments are modified. The modification method is: carry out end repair on the DNA fragments, Phosphorylate, add poly(A), and add sequencing adapters; then perform purification, PCR amplification, and construct a sequencin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com