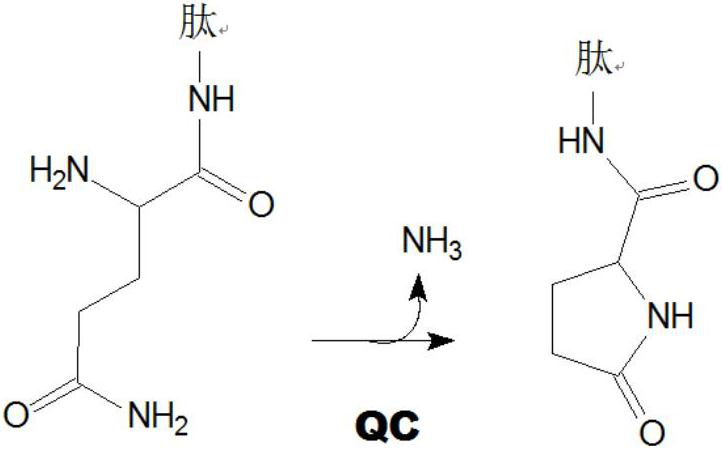
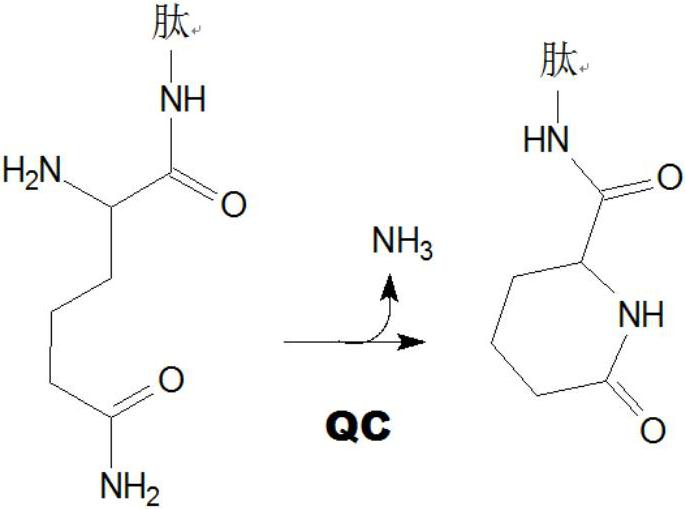
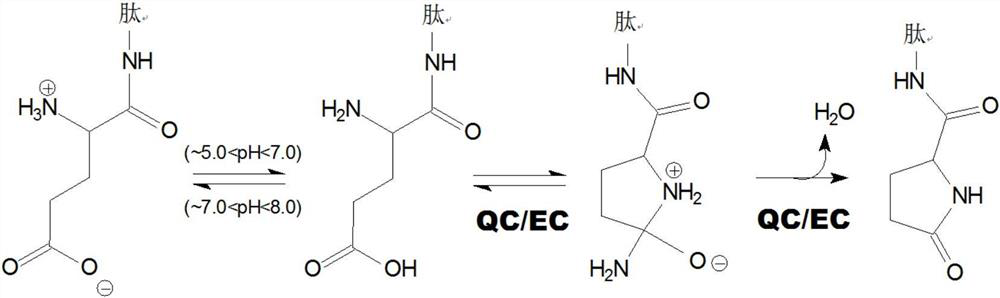
Inhibitors of glutaminyl cyclase
The technology of a compound, azetidine, is applied in the field of azetidinone derivatives, which can solve problems such as undisclosed sequence homology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0425] Compounds were synthesized according to method 1:
[0426] rac1-(1H-benzimidazol-5-yl)-4-phenylazetidin-2-one:
[0427] Step A: 5-aminobenzimidazole (1.33g, 10mmol), benzaldehyde (1.27g, 1.22mL, 12mmol) in 80mL of toluene gave 1.62g of crude product I;
[0428] Step B: I (0.433g, 2mmol), zinc powder (0.262g, 4mmol), TMS-Cl (0.180mL, 1.4mmol), ethyl bromoacetate (0.377mL, 3.4mmol). Yield: 0.089g (5.6%); MS m / z 285.1 (M+H) + ; 1 H NMR (DMSO, 400MHz): δ0.82-0.91 (m, H); 0.97-1.16 (m, 4H); 1.39-1.42 (m, H); 1.52-1.69 (m, 5H); 3.24-3.27 ( m, H); 3.42-3.46 (m, H); 4.48-4.52 (m, H); 6.92 (s, H); 7.56-7.59 (dd, H, 3 J=9.1Hz, 4 J=2.1Hz); 7.73-7.75 (d, H, 3 J=9.1Hz); 7.94-7.95 (d, H, 4 J=2.1Hz); 9.24(s, H), HPLC99%.
Embodiment 2 and 3
[0430] (R)-1-(1H-Benzo[d]imidazol-6-yl)-4-phenylazetidin-2-one 2
[0431]Example 1 (0.089 g) was subjected to chiral preparative HPLC separation to obtain compound 2. Yield: 0.025g, 1 H-NMR (DMSO-d6): δ12.25 (d, 1H); 8.13 (d, 1H); 7.53 (d, 1H); 7.46-7.30 (m, 6H); 7.12 (d, 1H); t, 1H); 3.63-3.57 (m, 1H); 2.92-2.86 (m, 1H); MS=264 (M+1); HPLC ~ 96.31%: Chiral HPLC ~ 92.48%
[0432] (S)-1-(1H-Benzo[d]imidazol-6-yl)-4-phenylazetidin-2-one 3
[0433] Example 1 (0.089 g) was subjected to chiral preparative HPLC to obtain compound 3. Yield: 0.025g, 1 H-NMR (DMSO-d6): δ12.25 (d, 1H); 8.13 (s, 1H); 7.46-7.30 (m, 7H); 7.12 (bs, 1H); 3.57(m, 1H); 2.90-2.86(m, 1H); MS=264(M+1); HPLC~99.00%: Chiral HPLC~98.35%.
Embodiment 4
[0435] Rac 1-(1H-benzo[d]imidazol-5-yl)-4-(2,6-difluoro-4-methoxyphenyl)azetidin-2-one 4
[0436] Step A: 5-aminobenzimidazole (0.690g, 5.2mmol), 2,5-difluoro-4methoxy-benzaldehyde (1.07g, 6.2mmol) in 80mL of toluene gave 1.66g of crude product I;
[0437] Step B: I (1.15 g, 4 mmol), zinc powder (0.524 g, 8.31 mmol), TMS-Cl (0.360 mL, 2.8 mmol), ethyl bromoacetate (0.754 mL, 6.8 mmol). Yield: 0.022g (1.2%); MS m / z 330.4 (M+H) + ; 1 H NMR (DMSO-d 6 , 400MHz): δ3.25-3.29(m, 1H), 3.63-3.67(m, 1H), 3.76(s, 3H), 5.49-5.51 8m, 1H), 6.80(d, 2H, 3J=11.2Hz) , 7.34 (dd, 1H, 3J = 8.8Hz, 4J = 2Hz), 7.50 (d, 1H, 4J = 2Hz), 7.74 (d, 1H, 3J = 8.8Hz), 9.10 (s, 1H), HPLC [A ]: 100%)
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com