Intron retention prediction model establishing method and prediction method thereof
A technology for predicting models and establishing methods, applied in biological neural network models, instruments, biological systems, etc., can solve the problems of restricting technology development, insufficient method robustness, and low method reliability, and achieve the effect of improving learning effects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
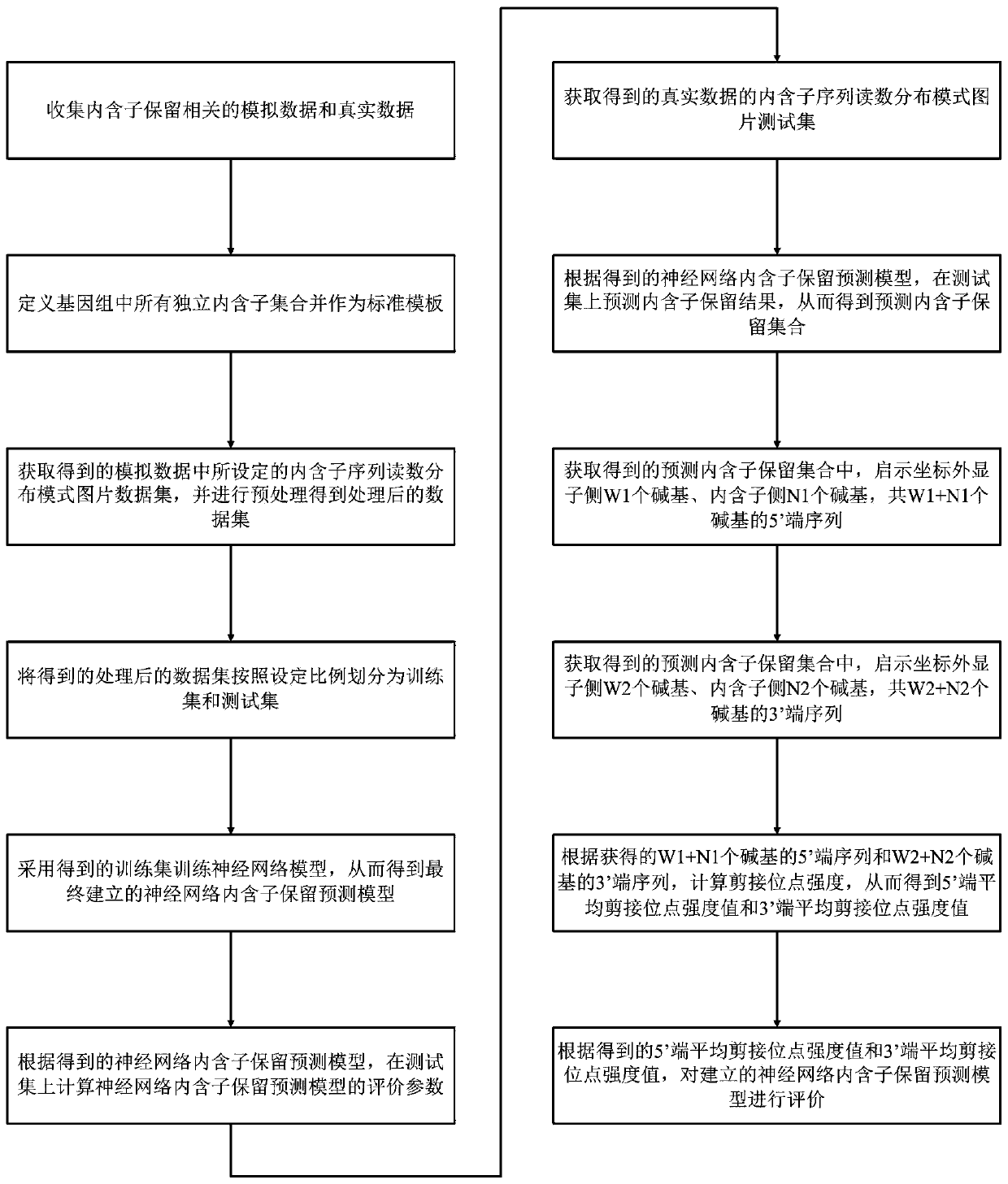
[0059] Such as figure 1 Shown is a schematic flow chart of the method for establishing the intron retention prediction model of the present invention: the method for establishing the intron retention prediction model provided by the present invention includes the following steps:
[0060] S1. Collect simulated data and real data related to intron retention; specifically, the BEER algorithm is used to generate a simulated data sequence file SIMU30 containing a determined number of introns; the sequencing depth of the simulated data sequence file SIMU30 is 30 million, and the number of reads The length is 100 bases, and it is set to generate 15,000 genes and 69,338 introns; and a real data sequence file APP from the Tau and APP mouse model research of the Alzheimer's Disease Accelerated Drug Cooperation Project, and the sequencing depth 100 million with a read length of 101 bases;
[0061] S2. Define all independent intron sets in the genome and use it as a standard template; t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Height | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com