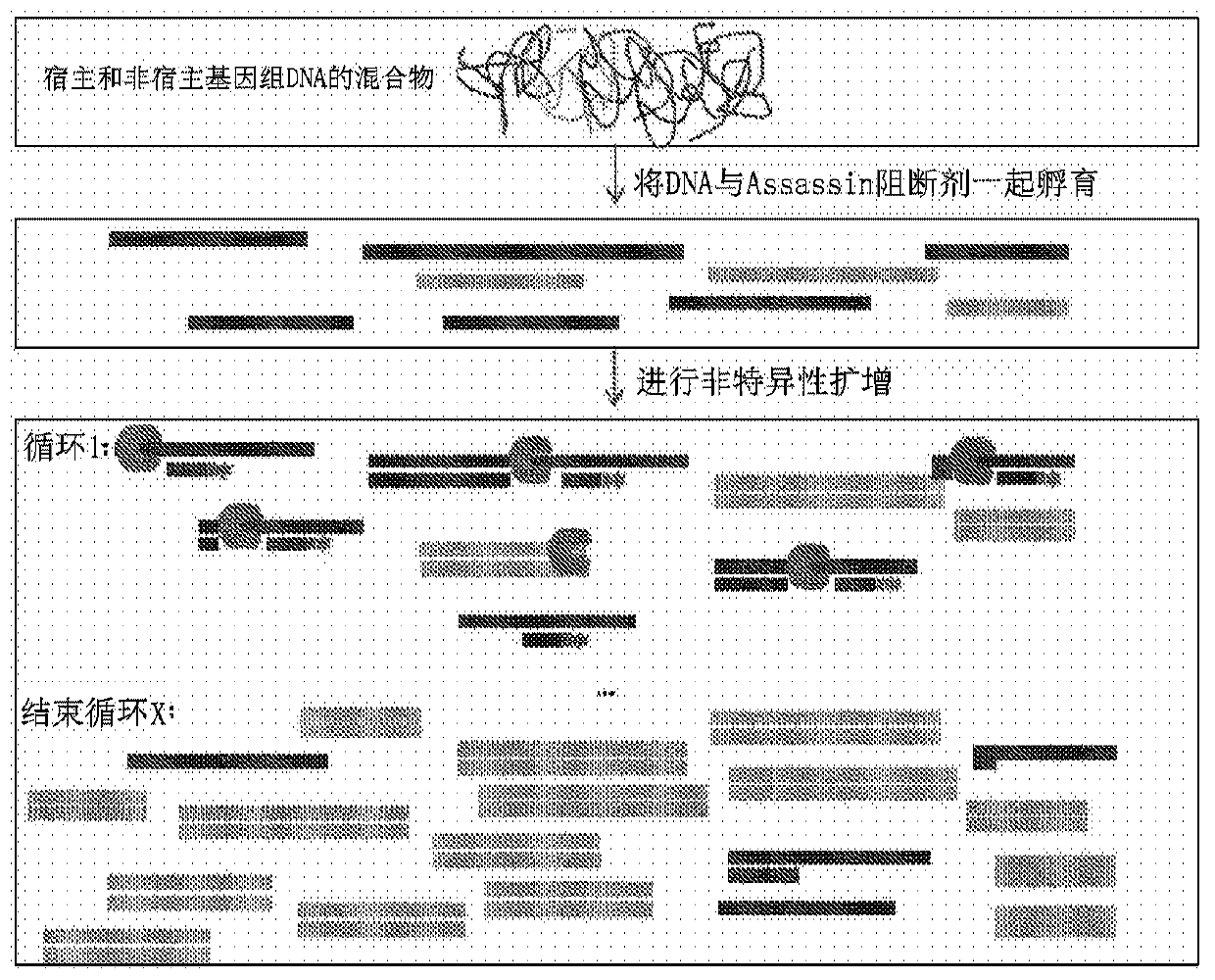
Selective enrichment of population of DNA in mixed DNA sample through targeted suppression of DNA amplification
A population and sample technology, applied in DNA/RNA fragments, recombinant DNA technology, microbial measurement/inspection, etc., can solve problems such as determining the resistance of complex polymorphisms
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0127] Design of Assassin Blocker DNA Oligonucleotide Sequence
[0128] In order to inhibit the amplification of a first population of DNA molecules comprising mammalian DNA sequences, Assassin blocker oligonucleotides were designed that are complementary to DNA sequences that are more abundant in mammalian DNA populations than microbial DNA populations appears more frequently in . The selected mammalian sequences are unique to the first population with minimal overlap with the second population of DNA to be amplified.
[0129] The human mitochondrial genome is small (approximately 16.6 kb) and contains sequences distributed on the plus and minus strands that are unique relative to both the human nuclear and bacterial genomes . The sequences of the Assassin blocker oligonucleotides are designed to be complementary to sequences that have a higher frequency per kilobase (kb) in mitochondrial genomes compared to bacterial, fungal, and viral genomes. Some non-limiting example...
Embodiment 2
[0168] Cross-linking of Assassin blockers to complementary sequences
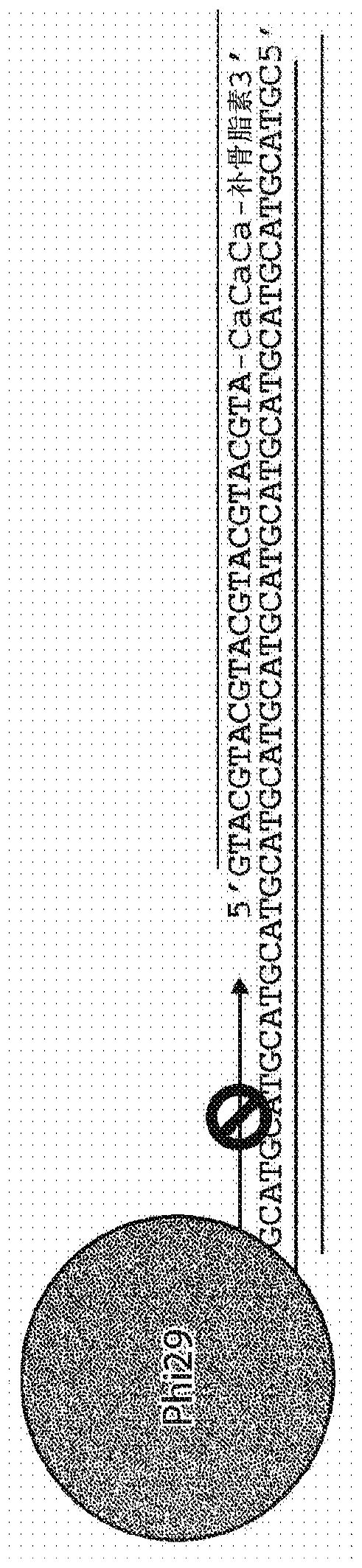
[0169] Amplification of the first population of human mitochondrial DNA can be blocked by cross-linking the Assassin blocker oligonucleotide designed in Example 1 with its complementary sequence in the first population of human mitochondrial DNA. This cross-linking blocks genome-wide amplification of sequences by strand-displacing polymerases such as phi29 ( figure 2 ).
[0170] Lyophilized Assassin blocker oligonucleotides with modified 3' amino groups were designed as described in Example 1 and purchased (Integrated DNA Technologies, Coralville, IA). Lyophilized oligonucleotides were resuspended in phosphate-buffered saline (1×PBS) and mixed with NHS-SPB (succinimidyl-[4-(psoralen-8-yloxy)]-butanol ester, "psoralen") molecules (ThermoFisher, Waltham, MA) were conjugated. The conjugation reaction was quenched with Tris-HCl and excess psoralen was removed with a desalting column (ThermoFisher, Wa...
Embodiment 3
[0172] Demonstration of sequence-specific covalent crosslinking of Assassin blockers to target DNA
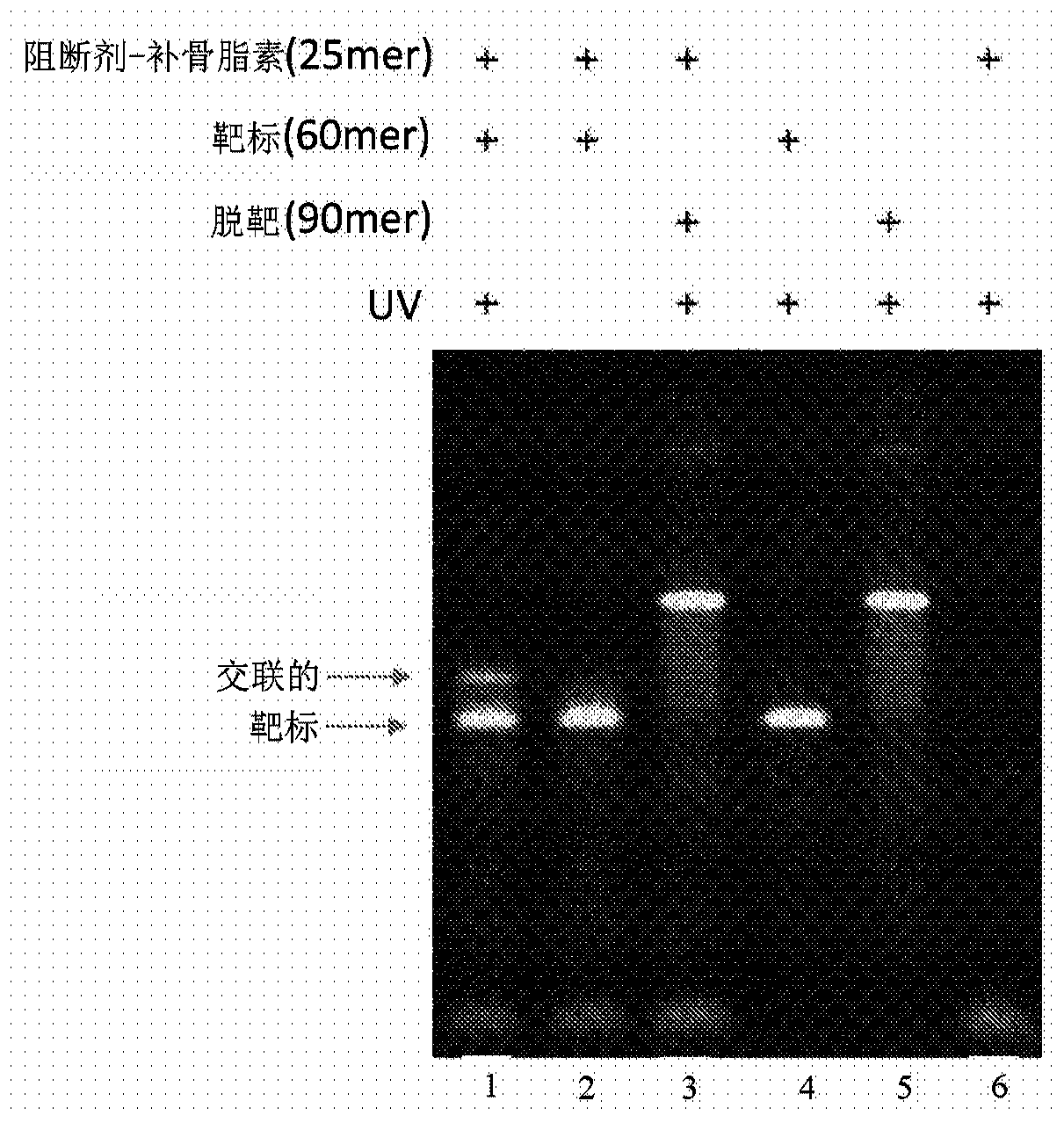
[0173] To demonstrate sequence-specific covalent crosslinking of Assassin blocker oligonucleotides to target DNA, different Assassin blocker oligonucleotides were conjugated to the 3' end with psoralen, target oligonucleotide The reaction mixture of acid and off-target oligonucleotides was added at 0.5 μM to a mixture of 40 mM Tris HCl (pH 7.9), 50 mM NaCl and 10 mM MgCl 2 Composition of the annealing buffer. The reaction mixture was heated to 94°C for 5 minutes and cooled to 16°C within 15 minutes. The reaction mixture was then treated with 350 nm ultra-violet (UV) light for 20 minutes. The reaction mixture products were separated on a 10% Tris-borate-EDTA(TBE)-urea gel in 1×TBE buffer. Gels were stained with SYBR-gold (ThermoFisher, Waltham, MA) and visualized on an E-gel Imager (ThermoFisher, Waltham, MA). ( image 3 ).
[0174] A reduction in human mitochondrial DNA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com