Circular RNA recognition method based on machine learning strategy
A technology of machine learning and recognition method, applied in the field of data science, can solve the problem of low recognition sensitivity and accuracy, and achieve the effect of cost saving
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
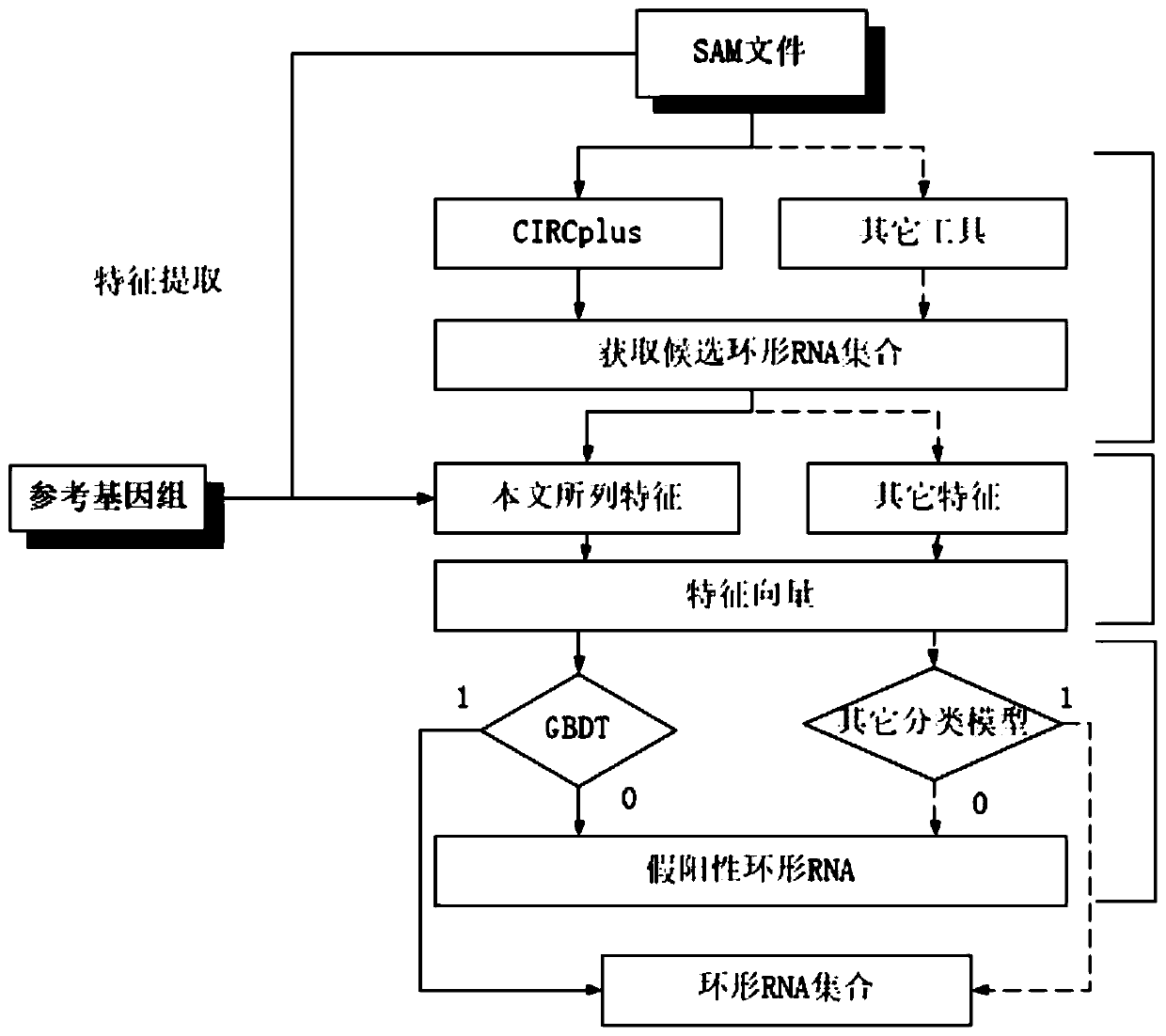
[0045] see figure 1 , the present invention a kind of circular RNA identification method based on machine learning strategy, comprises the following steps:
[0046] S1, input data
[0047] For the SAM file and gene annotation FASTA file generated by RNA-seq, the input data format requirements are: SAM format and FASTA format; run the existing circular RNA identification algorithm to obtain the candidate circular RNA set, and determine the breakpoint position of the candidate circular RNA;
[0048] Run the existing circular RNA identification algorithm to output a set of candidate circular RNAs, where the reference genome number is used, and the position of the 5' splicing site of the circular RNA is defined as the left breakpoint, using brk 1 Indicates; the position of the 3' splice site of circular RNA is defined as the right breakpoint, with brk 2 Represent; the two can jointly represent a candidate circular RNA; sort the circular RNAs in the candidate circular RNA set acc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com