MiRNA-disease association prediction method, system, terminal and storage medium
A prediction method and disease technology, applied in the field of bioinformatics, can solve problems such as inability to use association prediction, low prediction accuracy, and inability to fully characterize the complex relationship of miRNAs, and achieve high cost, high accuracy, and time-consuming problems. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
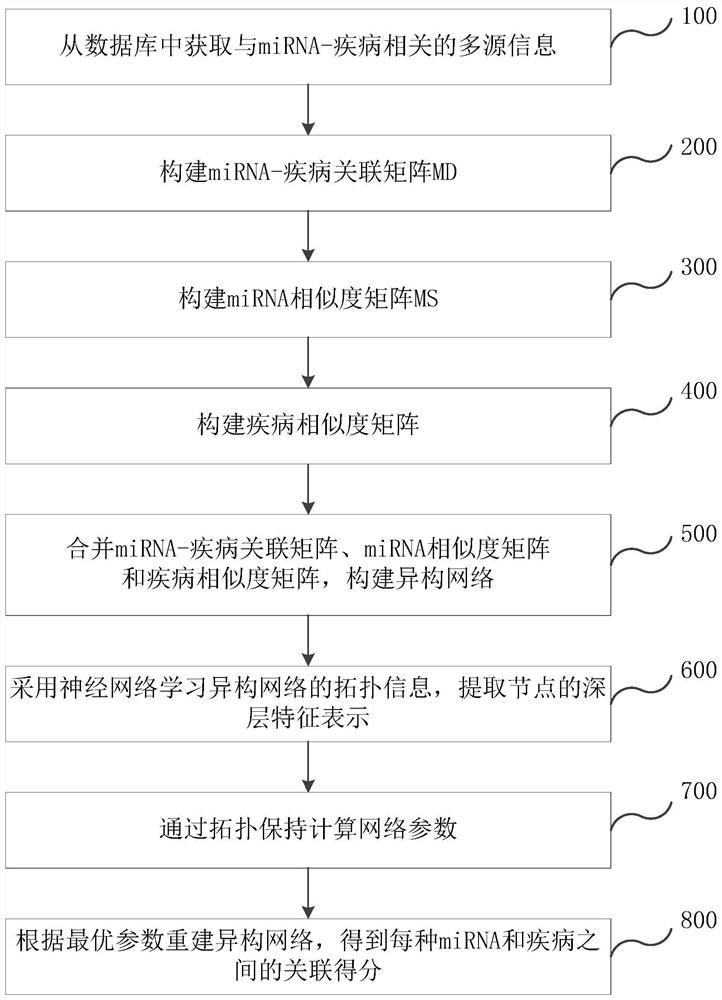
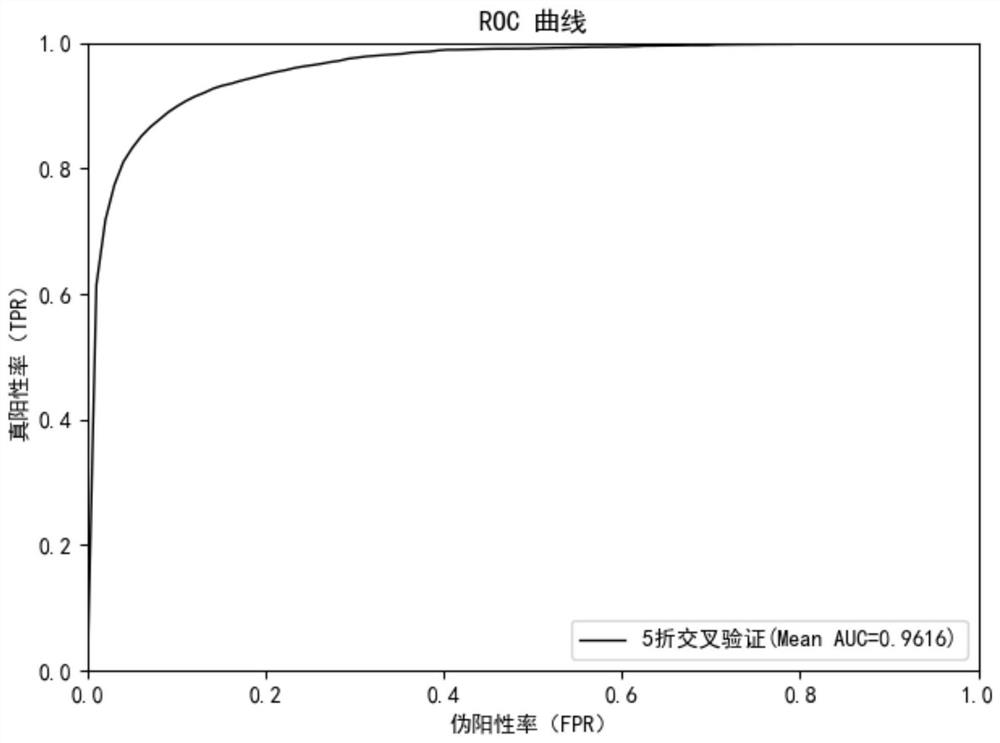
[0087] The following are the implementation steps of the miRNA-disease association prediction method according to the second embodiment of the application. The second embodiment of the application selects the disclosed data as test data, and uses a 5-fold cross-validation method to evaluate the technical solution of the application. Specifically:
[0088] S1: Obtain multi-source information related to miRNA-disease from the database;
[0089] S2: Construct miRNA-disease association matrix MD based on miRNA-disease association data;
[0090] In this step, after removing duplicate data from the test data, there are a total of 577 miRNAs and 336 diseases, with 6,441 associations. When constructing the miRNA-disease association matrix, all associations are randomly divided into 5 parts, of which 4 parts are used as the training set and 1 part is used as the test set. The association in the test set is set to 0 in the miRNA-disease association matrix, and the cycle is repeated 5 times. ...
Embodiment 2
[0100] In order to further evaluate the true performance of this application for predicting miRNA-disease associations, the following examples have conducted case studies on pancreatic cancer. In the case, all known associations related to pancreatic cancer are marked as unknown, and the model's ability to find these associated miRNAs is then studied. The top 50 miRNAs with the highest prediction scores are selected for verification in the dbDEMC2 and PhenomiR2 databases, which store the experimentally confirmed associations between miRNAs and diseases. It can be seen from the verification results that the 50 miRNAs (shown in Table 1) related to pancreatic cancer predicted by this application are all found in the database, which further proves the practicality of the model and the model can be used without any known Related miRNA predictions for related diseases.
[0101]
[0102]
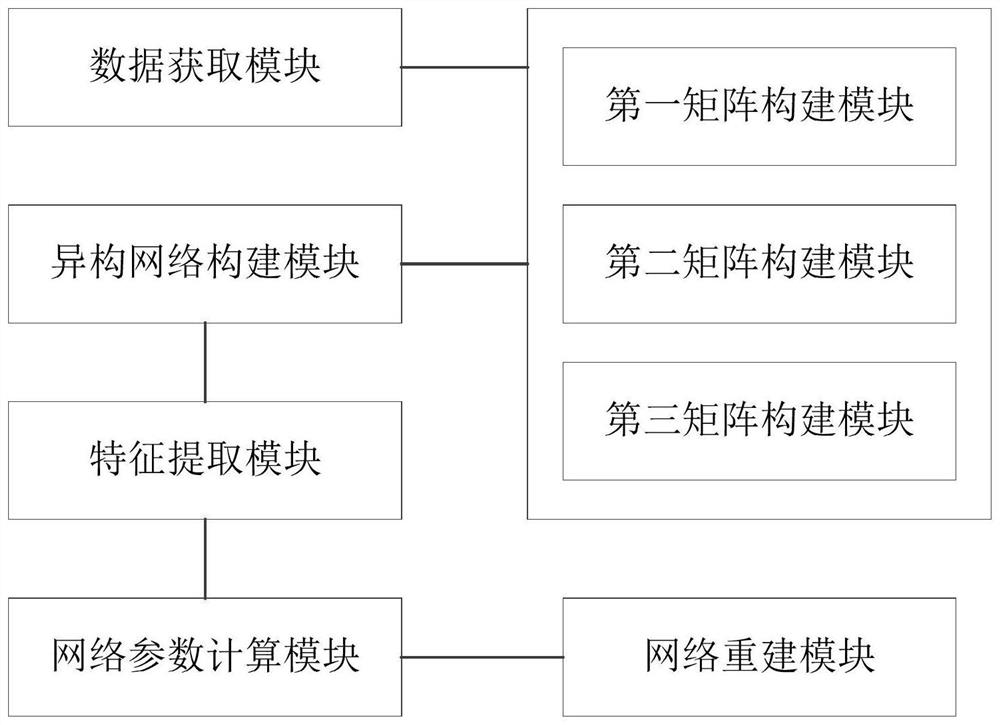
[0103] See image 3 , Is a schematic structural diagram of the miRNA-disease association predicti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com