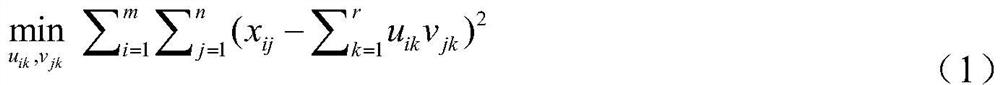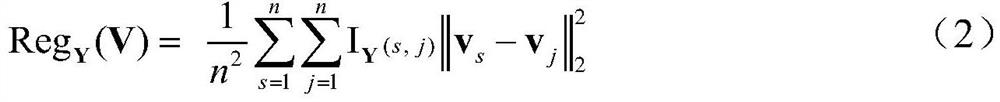Mutually-exclusively constrained graph Laplacian approach to heterogeneous cancer driver gene identification
A technology of driving genes and identification methods, applied in genomics, proteomics, medical automated diagnosis, etc., can solve problems affecting the effective identification of driving genes, and achieve the effect of improving the performance of gene identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0027] The present invention will be further described below in conjunction with the accompanying drawings and embodiments, and the present invention includes but not limited to the following embodiments.
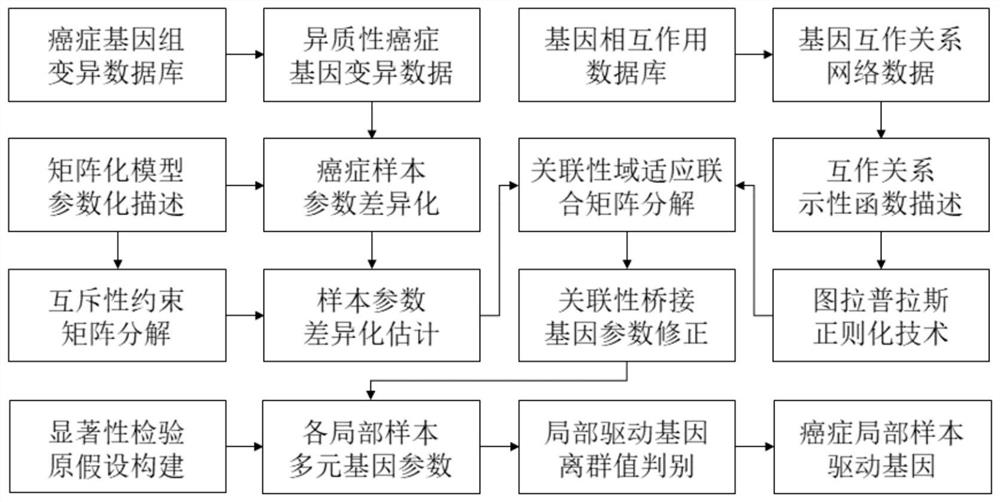
[0028] Such as figure 1 As shown, the present invention provides a method for identifying heterogeneous cancer driver genes with mutually exclusive constraint graph Laplacian, and its specific implementation process is as follows:
[0029] Step 1: Obtain cancer gene variation data through databases such as The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC). Data on gene interaction networks were collected from the STRING interaction database. The ID of the gene name was unified through the gene annotation database Database for Annotation, Visualization and Integrated Discovery (DAVID), so as to eliminate the phenomenon of homonym of genes in different sources of data.
[0030] Step 2: Use a matrix model to describe the heterogeneity of cancer...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com