Novel single-base editing technology and application thereof
A technology for editing enzymes and genes, which can be used in applications, recombinant DNA technology, genetic engineering, etc., and can solve the problems of large gene editing windows and low DNA editing accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0256] Example 1: Off-target RNA SNV detection for various single base editing systems
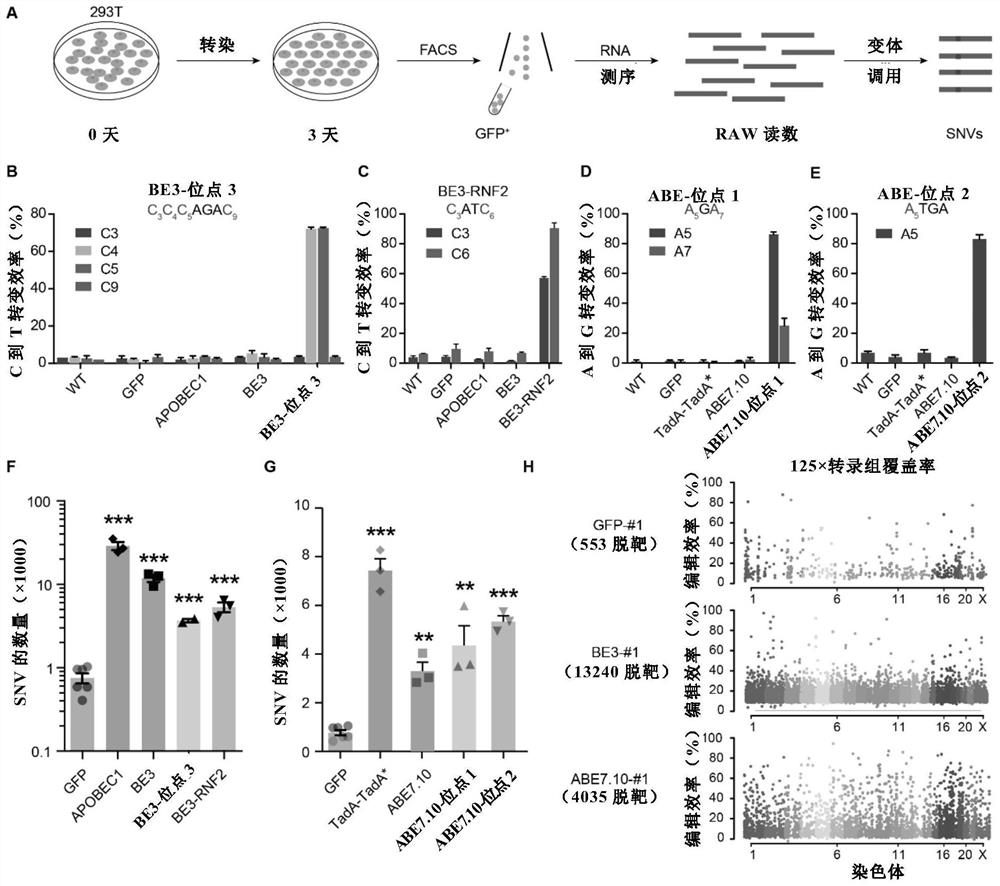
[0257] In this example, in order to assess the off-target effect of gene editing at the RNA level, CBE, BE3 (APOBEC1-nCas9-UGI) or ABE, ABE7.10 (TadA-TadA*-nCas9), and GFP with or without Single guide RNA (sgRNA) was transfected into cultured 293T cells. After 72 hours of incubation, GFP-expressing cells were harvested by FACS and then analyzed by RNA-seq. The experimental results of each group were compared with wild-type (WT, non-transfected) samples, calling RNA SNVs in each transfection group ( figure 1 A).
[0258] The 9 groups of transfected cells include expressing GFP, APOBEC1, BE3, BE3 with "site 3" sgRNA, BE3 with "RNF2" sgRNA, TadA-TadA*, ABE7.10, ABE7 with "site 1" sgRNA. 10. ABE7.10 cells with "site 2" sgRNA ( Figure 5 ).
[0259] First, the high targeting efficiency of DNA editing of BE3 and ABE7.10 in these 293T cells was verified using targeted deep sequencing, as sho...
Embodiment 2
[0263] Example 2: Characterization of off-target RNA SNVs
[0264] In this example, off-target RNA SNVs were characterized for each single base editing system.
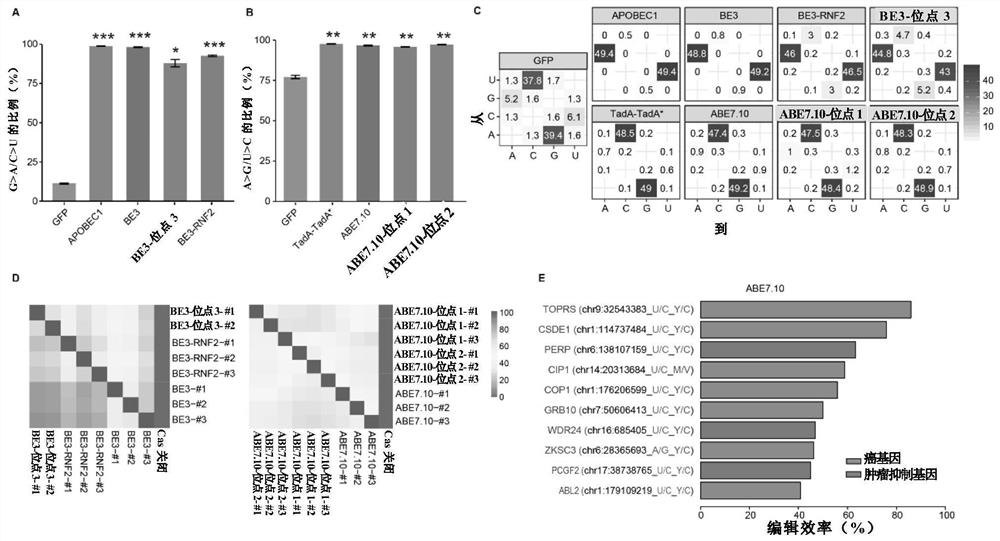
[0265] The result is as figure 2 with Figure 7-12 shown.
[0266] Notably, almost 100% of the RNA SNVs identified in BE3-treated cells were G to A or C to U mutations, significantly higher than in GFP-transfected cells (e.g. figure 2 A and 2C and Figure 7 ). This mutational bias is identical to APOBEC1 itself, suggesting that these mutations are not spontaneous but induced by BE3 or APOBEC1.
[0267] Correspondingly, 95% of ABE7.10-induced mutations were A to G or U to C, consistent with the effect of ABE7.10 (eg figure 2 B and 2C and Figure 7 ).
[0268] From the results, it can also be noticed that the GFP group also exhibits some bias for A to G and U to C mutations (eg figure 2 C), which may be due to an innate mutation preference.
[0269] In any two samples of BE3- or ABE7.10-transfection groups...
Embodiment 3
[0272] Example 3: Single-cell RNA SNV analysis of cells transfected with a single base editing system
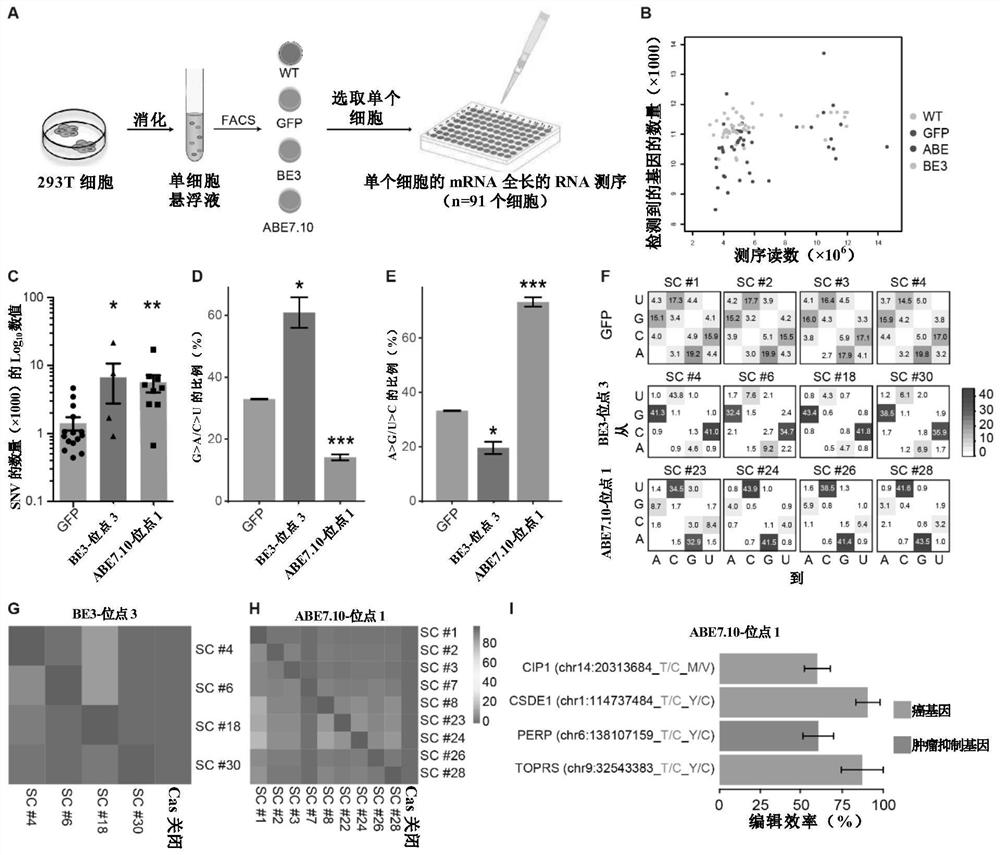
[0273] In this example, single-cell RNA-seq sequencing was performed on four groups of cells (WT, GFP, BE3-site 3 and ABE7.10-site 1) to avoid random off-target signal loss due to population averaging .
[0274] The result is as image 3 with Figure 13-17 shown.
[0275] On average, 10,932 RefSeq genes were detected in each single cell by approximately 6.07 million sequencing reads, with results such as image 3 Shown in B. Cells with high expression levels of the indicated deaminases were selected for further analysis, and the results Figure 13 shown. Also, severe RNA off-targets and similar mutational patterns were observed in those cells expressing base edits (e.g. image 3 C to 3F and Figure 14 with 15 ).
[0276] Interestingly, the percentage of off-target sites shared by any BE3- or ABE7.10-edited cells (4.5+ / -1.0%) was much lower than that of the cell pop...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com