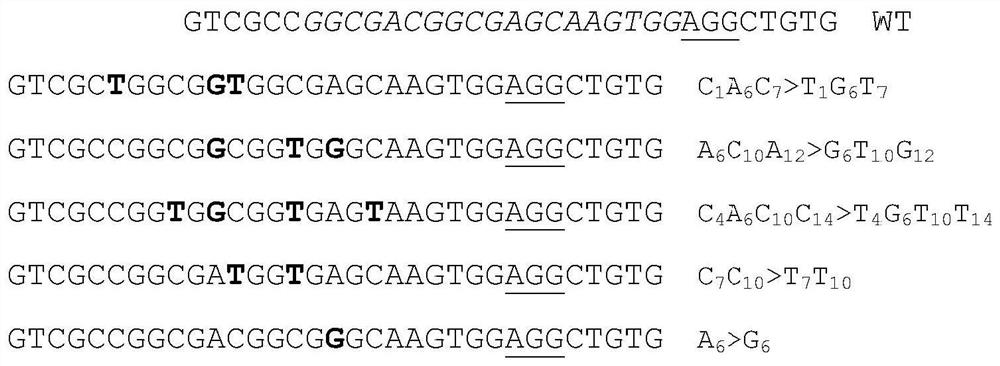Co-transcription unit gene ABE-CBE system for bidirectional single base editing of rice and application thereof
An ABE-CBE, single-base technology, applied in the field of biotechnology and plant genetic engineering, can solve the problems of fixed-point modification dependence, low efficiency, and the ability to edit genes accurately, so as to achieve wide application value and reduce unexpected bases The effect of replacing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0119] Example 1 - Splicing of the mutant TadA-nSpCas9-LjCDAL1 gene
[0120] The gene of the present application is named mutant TadA-nSpCas9-LjCDAL1, and its sequence is shown in SEQ ID NO:1.
[0121] First, through a large number of experiments, the applicant designed and screened a lamprey-like cytosine deaminase (cytidine deaminase, LjCDAL1) gene with a wide window and high editing efficiency, named Os-LjCDAL1 (sequence such as SEQ ID NO: 2 ), its editing efficiency can reach more than 80%. After the Os-LjCDAL1 gene was sent to Suzhou Jinweizhi Biotechnology Co., Ltd. for synthesis, it was connected to the PUC57-AMP vector to form the PUC57-AMP-LjCDAL1 vector, and loaded into E. coli XL-blue strain.
[0122] Secondly, the mutant adenine deaminase TadA suitable for rice was artificially synthesized, named mutant TadA, connected to the PUC57-AMP vector to form the PUC57-AMP-mutant TadA vector, and loaded into E. coli XL-blue strain .
[0123] According to the Gibson splic...
Embodiment 2
[0133] Example 2—Construction of plant targeting vector containing mutant TadA-nSpCas9-LjCDAL1 gene
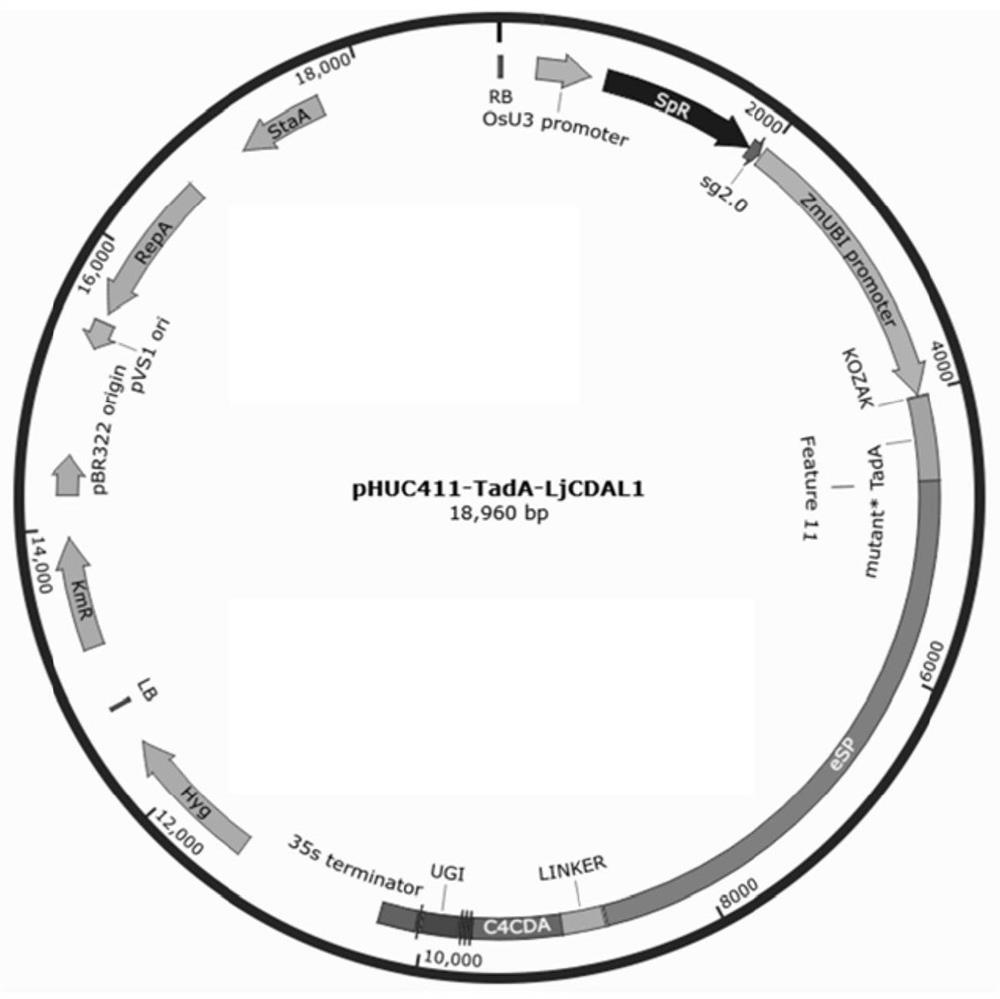
[0134] From Escherichia coli XL-blue containing the mutant TadA-nSpCas9-LjCDAL1 vector, use the Axygen plasmid extraction kit to extract the plasmid, digest it with NotI / SacI, and recover the OsSpCas9-LjCDAL1 fragment. At the same time, pHUC411 was linearized using NotI / SacI enzymes, pHUC411 was recovered, and the above-mentioned mutant TadA-nSpCas9-LjCDAL1 fragment and pHUC411 fragment were connected with T4 ligase (purchased from TaKaRa Company) to obtain the plant expression vector pHUC411mutant TadA-nSpCas9 -LjCDAL1( figure 1 ), named pHUC411-TadA-LjCDAL1.
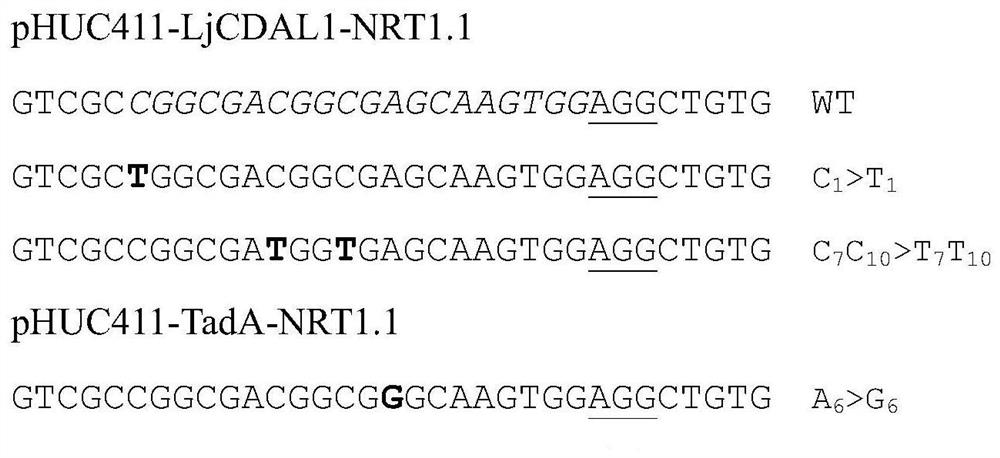
[0135] Select the nucleotide sequence CGGCGACGGCGAGCAAGTGG of position 3105-3124 in rice NRT1.1 gene (Os10g0554200) AGG , (the underlined part is the PAM sequence of the 5'NGG-3' structure), as the targeting site. The target site sequence was fused to pHUC411-TadA-LjCDAL1 to form pHUC411-TadA-LjCDAL1-NRT1.1. The p...
Embodiment 3
[0136] Example 3—Genetic transformation of rice using pHUC411-TadA-LjCDAL1-NRT1.1 as a targeting vector and acquisition of mutants.
[0137] 1. Induction and pre-culture of mature embryo callus
[0138] The mature seeds of Nipponbare are shelled, and the seeds with normal appearance and cleanness without mildew are selected, shaken for 90 sec with 70% alcohol, and poured off the alcohol; Add 1 drop of Tween20) solution to 1 ml to wash the seeds, and shake on the shaker for 45min (180r / min). Pour off the sodium hypochlorite, wash with sterile water 5-10 times until there is no smell of sodium hypochlorite, finally add sterile water, soak overnight at 30°C. Use a scalpel to separate the embryos along the aleurone layer, put the scutellum up on the induction medium (see Table 1 for ingredients), 12 embryos / dish, and culture in the dark at 30°C to induce callus.
[0139] Two weeks later, spherical, rough, light yellow secondary callus appeared, and pre-cultivation operation coul...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com