Drug-target interaction prediction method based on hypergraph neural network
A neural network and prediction method technology, applied in the field of drug-target interaction prediction based on hypergraph neural network, can solve problems such as difficult to capture high-order complex relationships between drugs and targets, and achieve good prediction results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
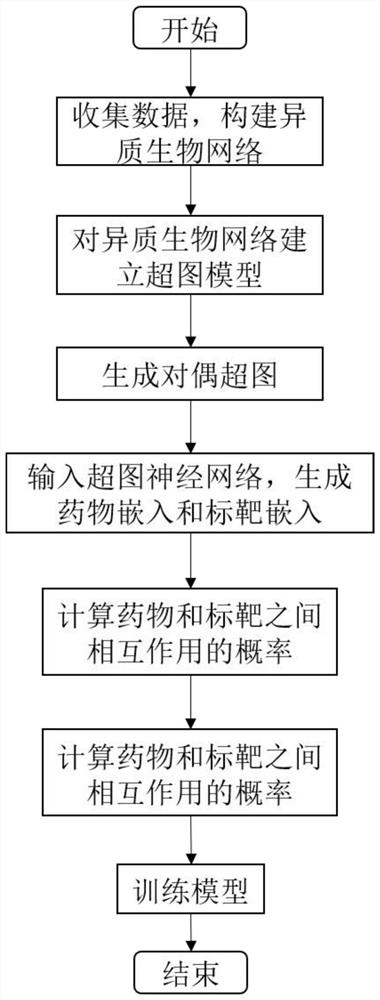
[0054] The drug discovery method based on hypergraph neural network proposed by the present invention is implemented according to the following steps.
[0055] Download the approved Target Drug-UniprotLinks from the public database Drugbank version (5.1.7). This contains 2141 drugs, 2635 targets and 11022 drug-target interactions. Divide it into training set and test set.
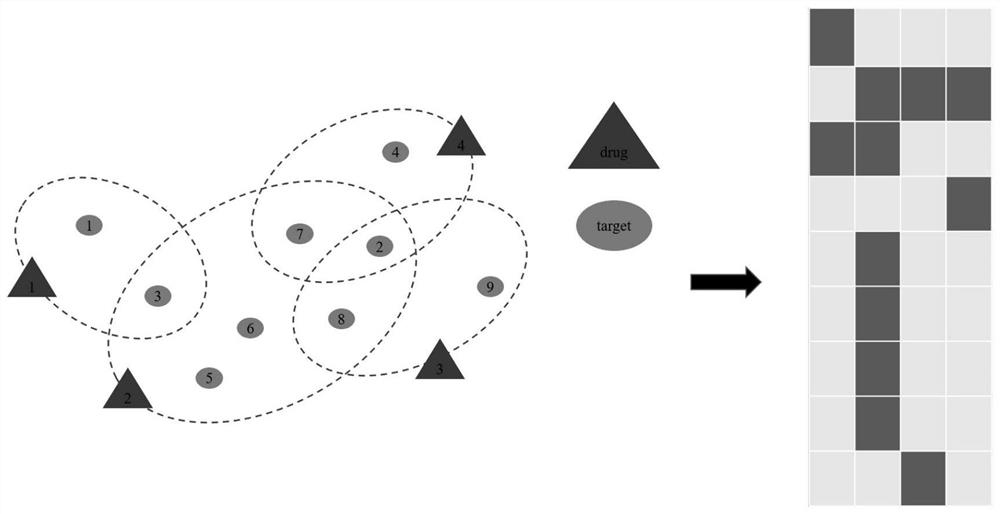
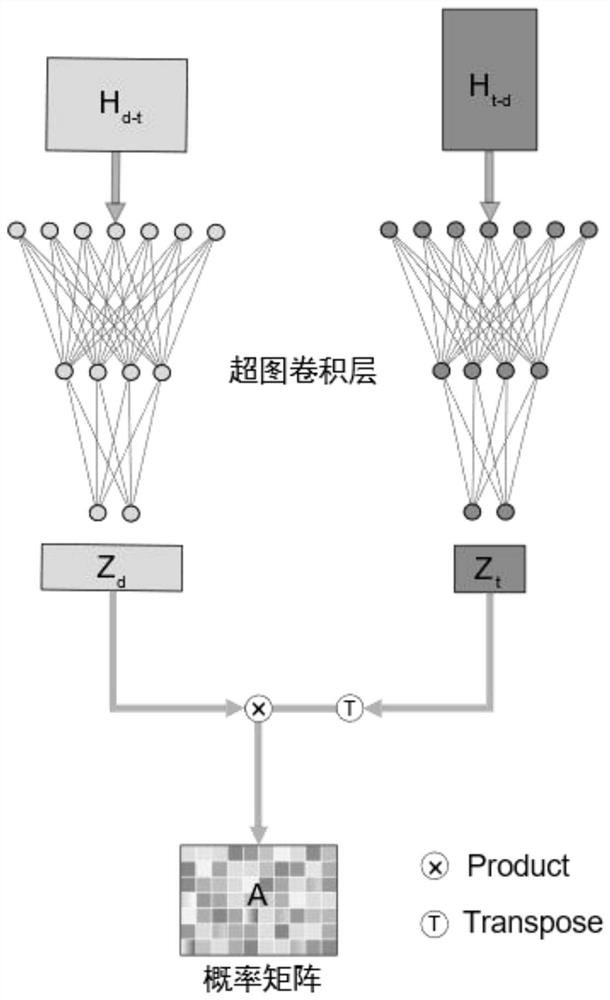
[0056] Model the heterogeneous biological network composed of two heterogeneous nodes in the training set to obtain the heterogeneous biological hypergraph G={V={v 1 ,...,v M},E={e 1 ,...,e N}}. The hypergraph is different from the traditional graph model. An edge in the hypergraph can no longer only connect two nodes, but can connect more than two nodes, which is called a hyperedge. Specifically, a drug can use a hyperedge to link multiple targets; a target can also use a hyperedge to link multiple drugs. Such as figure 1 As shown, using the incidence matrix Represents the high-order complex rela...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com