Simulation analysis method of DNA polyhedron with special branch number
A technology of simulation analysis and polyhedron, which is applied in the field of simulation analysis of DNA polyhedron, can solve the problems of inability to judge the influence of DNA polyhedron properties, lack of research on the structure of DNA polyhedron, etc., and achieve the effect of low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
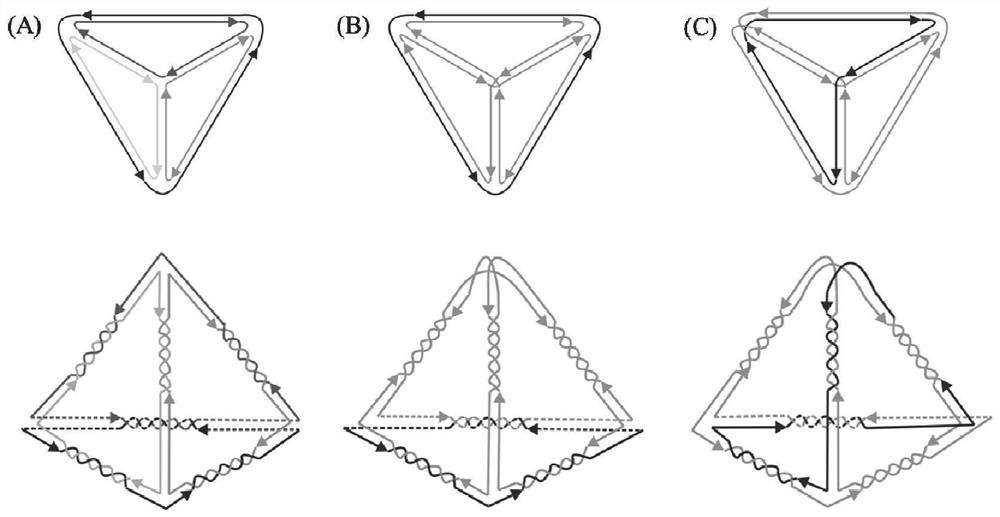
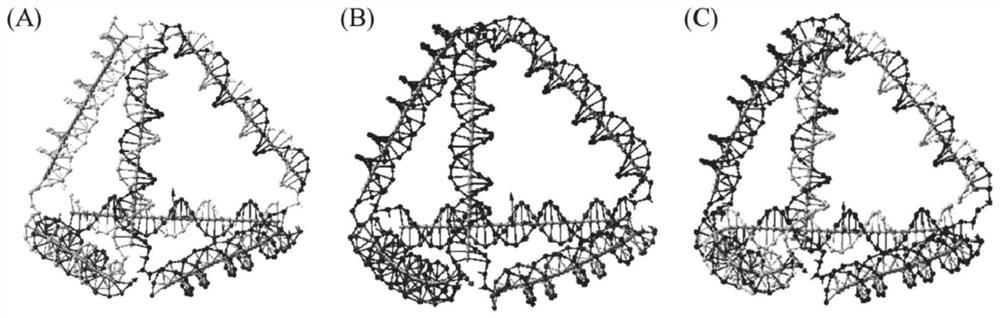
[0068] (1) Firstly, draw a four-branched DNA tetrahedral link diagram. The number of branches of the DNA tetrahedron is set to 4, the number of intersections on each side is set to 6, the length of the double helix side is set to 31nt, and the length of the linker at the apex is set to 3T. The link graph of DNA polyhedron structure is as follows figure 1 As shown, the whole structure is composed of 4 DNA single strands, and the length of each strand is 3 tetrahedral side lengths, and the DNA tetrahedron is named 4s.
[0069] (2) Use the software uniquimer3D to design the sequence of the DNA polyhedron. The sequence of the DNA polyhedron is as follows, where the underlined part represents the linker:
[0070] strand 1:
[0071] AAACTACTCCTCGAAGTGATTTGTACCGTCT TTT GATAGGGCGGGACCCGG
[0072] GATAGCATATGGGT TTT TGCCCGGATCGAGACCCCTCAATTCGGGAGG
[0073] strand 2:
[0074] ACCCATATGCTATCCCGGGTCCCGCCCTATC TTT ATTTGCGTGATCGCATCACTACCAGACGGAC TTT TAAAAGGGGAATCCCTGCCACGTGAAT...
Embodiment 2
[0087] (1) Firstly, draw a two-branched DNA tetrahedral link graph. The number of branches of the DNA tetrahedron is set to 2, the number of intersections on each side is set to 6, the length of the double helix side is set to 31nt, and the length of the linker at the apex is set to 3T. The link graph of DNA polyhedron structure is as follows figure 1 As shown, the whole structure is composed of 2 DNA single strands, one of which has a length of 3 tetrahedral sides, and the other has a length of 9 tetrahedral sides. The DNA tetrahedron is named 2s1.
[0088] (2) Use the software uniquimer3D to design the sequence of the DNA polyhedron. The sequence of the DNA polyhedron is as follows, where the underlined part represents the linker:
[0089] strand 1:
[0090] ATCGTCTATAGTAAGTTTTTTCCTAACGCAGG TTT TGTTTTCGCGTTACTTTATAGCGGATTTTCA TTT TTGGATCAAATATGAGTAGGTCACGTATCTA TTT TCGGATCCTAGGCTCAGGATCTGGGTATCCA TTT TAGCACATTCAAATCTCCGTTCAGGGGCTCGG TTT TGAAAATCCGCTATAAAGTAACGCGA...
Embodiment 3
[0101] (1) Firstly, draw a two-branched DNA tetrahedral link graph. The number of branches of the DNA tetrahedron is set to 2, the number of intersections on each side is set to 6, the length of the double helix side is set to 31nt, and the length of the linker at the apex is set to 3T. The link graph of DNA polyhedron structure is as follows figure 1 As shown, the whole structure is composed of 2 DNA single strands, one of which has a length of 4 tetrahedral sides, and the other has a length of 8 tetrahedral sides. The DNA tetrahedron is named 2s2.
[0102] (2) Use the software uniquimer3D to design the sequence of the DNA polyhedron. The sequence of the DNA polyhedron is as follows, where the underlined part represents the linker:
[0103] strand 1:
[0104] GGTCGCTGTCGAAAGGCAGTTTCCTAGCAAT TTT TTCGCACGGTGGAGAGTCCGTCTTAACCGCC TTT TGCCGTCCGACTGGATGTTCAGTTCCTCAAAA TTT GCTGTGTAGGTCTGACGCAAAGATCGTACAT TTT ATTGCTAGGAAACTGCCTTTCGACAGCGACC TTT GGGTTTTGCCCTTGTTCAGGCCATGCA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com